Abstract
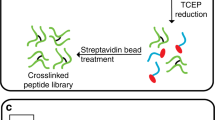
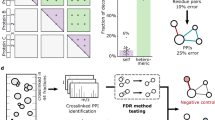
We have developed pLink, software for data analysis of cross-linked proteins coupled with mass-spectrometry analysis. pLink reliably estimates false discovery rate in cross-link identification and is compatible with multiple homo- or hetero-bifunctional cross-linkers. We validated the program with proteins of known structures, and we further tested it on protein complexes, crude immunoprecipitates and whole-cell lysates. We show that it is a robust tool for protein-structure and protein-protein–interaction studies.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Singh, P., Panchaud, A. & Goodlett, D.R. Anal. Chem. 82, 2636–2642 (2010).
Zhang, H. et al. Mol. Cell. Proteomics 8, 409–420 (2009).
Petrotchenko, E.V., Serpa, J.J. & Borchers, C.H. Mol. Cell. Proteomics 10, M110.001420 (2011).
Kao, A. et al. Mol. Cell. Proteomics 10, M110.002212 (2011).
Hermanson, G.T. Bioconjugate Techniques 2nd edn Ch. 4, 241–243 (Academic, London, 2008).
Li, S. et al. Genes Dev. 25, 2409–2421 (2011).
Dragon, F. et al. Nature 417, 967–970 (2002).
Grandi, P. et al. Mol. Cell 10, 105–115 (2002).
Krogan, N.J. et al. Mol. Cell 13, 225–239 (2004).
Champion, E.A., Lane, B.H., Jackrel, M.E., Regan, L. & Baserga, S.J. Mol. Cell. Biol. 28, 6547–6556 (2008).
Lee, L.W., Lo, H.W. & Lo, S.J. Gene 455, 16–21 (2010).
Aittaleb, M. et al. Nat. Struct. Biol. 10, 256–263 (2003).
Lin, J. et al. Nature 469, 559–563 (2011).
Rinner, O. et al. Nat. Methods 5, 315–318 (2008).
Xu, H., Hsu, P.H., Zhang, L., Tsai, M.D. & Freitas, M.A. J. Proteome Res. 9, 3384–3393 (2010).
Su, C. et al. Nucleic Acids Res. 36, D632–D636 (2008).
Kubala, M.H., Kovtun, O., Alexandrov, K. & Collins, B.M. Protein Sci. 19, 2389–2401 (2010).
Panchaud, A., Singh, P., Shaffer, S.A. & Goodlett, D.R. J. Proteome Res. 9, 2508–2515 (2010).
Brenner, S. Genetics 77, 71–94 (1974).
Acknowledgements
The authors wish to express gratitude to A.F. Hühmer and D. Horn (Thermo Fisher Scientific) for discussion; L.-L. Du (National Institute of Biological Sciences, Beijing (NIBS)) for critical suggestions; S.J. Lo (Chang Gung University) for the fib-1∷GFP strain; E.-Z. Shen, H.-Q. Wang, X.-D. He and G.-H. Cai (NIBS) for help with microscopy, GFP immunoprecipitation and peptide experiments; and R. Aebersold and T. Walzthöni (ETH Zurich) for help with xQuest search. We thank Z. Yuan, C. Liu, R.-X. Sun, Y. Fu and other members of the pFind team for discussion and support. This work was funded by the Ministry of Science and Technology of China (863 grant 2007AA02Z1A7 and 973 grant 2010CB835203 to M.-Q.D.; 863 grant 2008AA022310 and 973 grant 2010CB835402 to K.Y.; 973 grants 2012CB910602 and 2010CB912701 to S.-M.H.), the CAS Knowledge Innovation Program (KGCX1-YW-13) and an ICT basic research grant to S.-M.H. We thank the municipal government of Beijing for funds allocated to NIBS.
Author information
Authors and Affiliations
Contributions
B.Y. performed most wet-lab experiments and data analysis, and contributed to manuscript preparation; Y.-J.W. developed pLink; M.Z. performed peptide experiments (with B.Y.), non-BS3 cross-linking experiments and Y2H experiments; S.-B.F. fixed software bugs and maintained the system; J.L., S.L. and S.-K.L. purified the UTP-B, CNGP and F15E11.13-F15E11.14 protein complexes, respectively; K.Z. and L.-Y.X. developed pLabel for cross-links; H.C., H.-F.C. and L.-H.W. contributed to pLink development; Y.-X.L. contributed to data analysis; Y.-H.D. contributed to non-BS3 cross-linking experiments; Z.H. and S.C. contributed to MS analysis; K.Y. provided crucial samples and contributed to data interpretation and manuscript preparation; S.-M.H. directed software development and revised the manuscript; M.-Q.D. conceived the study, directed wet-lab experiments and wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–18, Supplementary Tables 1–20 and Supplementary Note (PDF 4178 kb)
Rights and permissions
About this article
Cite this article
Yang, B., Wu, YJ., Zhu, M. et al. Identification of cross-linked peptides from complex samples. Nat Methods 9, 904–906 (2012). https://doi.org/10.1038/nmeth.2099
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nmeth.2099
This article is cited by
-
Diverse modes of H3K36me3-guided nucleosomal deacetylation by Rpd3S
Nature (2023)
-
Structural basis of nucleosome deacetylation and DNA linker tightening by Rpd3S histone deacetylase complex
Cell Research (2023)
-
Characterization of protein unfolding by fast cross-linking mass spectrometry using di-ortho-phthalaldehyde cross-linkers
Nature Communications (2022)
-
HRS phosphorylation drives immunosuppressive exosome secretion and restricts CD8+ T-cell infiltration into tumors
Nature Communications (2022)
-
Structural and functional characterization of the Spo11 core complex
Nature Structural & Molecular Biology (2021)