As more genomes are assembled from scratch, scientists are struggling to assess and improve their quality.
Main
Asked how mature the field of genome assembly is, Ian Korf at the University of California, Davis, compares it to a teenager with great capabilities. “It's got bold assertions about what it can do, but at the same time it's making embarrassing mistakes,” he says. Perhaps the biggest barrier to maturity is that there are few ways to distinguish true insight from foolish gaffe. When a species' genome is newly assembled, no one knows what's real, what's missing, and what's experimental artifact.
That's not slowing the pace of assembly. In 2009, entomologists launched an initiative to sequence 5,000 insect genomes in a project known as i5k. Shortly thereafter, an international collaboration called Genome 10K organized around a goal to sequence thousands of vertebrate species. As of February 2012, The Genomes OnLine Database1 hosted by the Joint Genome Institute of the US Department of Energy, in Walnut Creek, California, listed over 15,000 sequencing projects; some 12,000 of these are still in progress or in planning stages. The Shenzhen, China–based global sequencing center BGI has set itself a goal of sequencing a million human genomes, a million microbe genomes and another million plant and animal genomes. Researchers are even eager to do de novo assembly on human genomes, the better to discover variation that is hidden when sequencing data are aligned to a reference.

Dozens of computer programs have been written to turn raw sequencing data into intact assemblies. But despite the amount of human, computing and laboratory resources poured into assembly, key questions remain unanswered. What combination of sequencing data and computer algorithms can produce the highest-quality assembly? More fundamentally, once a genome is assembled, how can scientists tell how good it is?
Millions of pieces with multiple copies
As genome assembly programs stitch together an organism's chromosomes from fragmented reads of DNA, they perform some of the most complex computations in all of biology. Sanger sequencing, the first mainstream sequencing technology, produces DNA fragments of up to 1,000 base pairs; adjacent reads usually overlap by a couple of hundred base pairs. This essentially turns the haploid human genome into a blank 30-million-piece jigsaw puzzle, complicated by the facts that some pieces are missing altogether and some pieces contain errors. To compensate, assemblers need about eight copies of each piece of the genome.
Short-read sequencing technologies have made the computational challenge harder. Next-generation sequencers can read base pairs at a hundredth to a thousandth of the cost of Sanger sequencing, but the reads are much shorter. With short-read sequencing technologies, the human-genome puzzle could contain 2 or 3 billion pieces with 100 copies of each piece.
Errors in assembly occur for many reasons. Pieces are often incorrectly discarded as mistakes or repeats; others are joined up in the wrong places or orientations. Researchers will be grappling with these kinds of issues for a while, says Adam Felsenfeld, director of the Large-Scale Sequencing program at the National Human Genome Research Institute (NHGRI), in Bethesda, Maryland. “Very long, very high-quality reads will do wonders for assembly, and fix many of these issues,” he says. “But we are not there yet.”
To assemble a genome, computer programs typically use data consisting of single and paired reads. Single reads are simply the short sequenced fragments themselves; they can be joined up through overlapping regions into a continuous sequence known as a 'contig'. Repetitive sequences, polymorphisms, missing data and mistakes eventually limit the length of the contigs that assemblers can build.
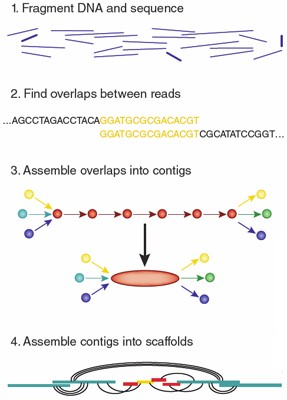
Paired reads typically are about the same length as single reads, but they come from either end of DNA fragments that are too long to be sequenced straight through. Depending on the preparation technique, that distance can be as short as 200 base pairs or as large as several tens of kilobases. Knowing that paired reads were generated from the same piece of DNA can help link contigs into 'scaffolds', ordered assemblies of contigs with gaps in between. Paired-read data can also indicate the size of repetitive regions and how far apart contigs are.
Assessing quality is made more difficult because sequencing technology changes so quickly. In January of this year, Life Technologies launched new versions of its Ion Torrent machines, which can purportedly sequence a human genome in a day, for $1,000 in equipment and reagents. In February, Oxford Nanopore Technologies announced a technology that sequences tens of kilobases in continuous stretches, which would allow genome assembly with much more precision and drastically less computational work. Other companies, such as Pacific Biosciences, also have machines that produce long reads, and at least some researchers are already combining data types to glean the advantages of each.
Software engineers who write assembly programs know they need to adapt. “Every time the data changes, it's a new problem,” says David Jaffe, who works on genome assembly methods at the Broad Institute in Cambridge, Massachusetts. “Assemblers are always trying to catch up to the data.” Of course, until a technology has been available for a while, it is hard to know how much researchers will use it. Cost, ease of use, error rates and reliability are hard to assess before a wider community gains more experience with new procedures. Luckily, ongoing efforts for evaluating short-read assemblies should make innovations easier to evaluate and incorporate.
Judging genomes
In the absence of a high-quality reference genome, new genome assemblies are often evaluated on the basis of the number of scaffolds and contigs required to represent the genome, the proportion of reads that can be assembled, the absolute length of contigs and scaffolds, and the length of contigs and scaffolds relative to the size of the genome.
The most commonly used metric is N50, the smallest scaffold or contig above which 50% of an assembly would be represented. But this metric may not accurately reflect the quality of an assembly. An early assembly of the sea squirt Ciona intestinalis had an N50 of 234 kilobases. A subsequent assembly extended the N50 more than tenfold, but an analysis by Korf and colleagues showed that this assembly lacked several conserved genes, perhaps because algorithms discarded repetitive sequences2. This is not an isolated example: the same analysis found that an assembly of the chicken genome lacks 36 genes that are conserved across yeast, plants and other organisms. But these genes seem to be missing from the assembly rather than the organism: focused re-analysis of the raw data found most of these genes in sequences that had not been included in the assembly.
Though the sea squirt and chicken genomes were assembled several years ago, such examples are still relevant because assembly is more difficult with the shorter reads used today, says Deanna Church, a staff scientist at the US National Institutes of Health who leads efforts to improve assemblies for the mouse and human genomes. “In my experience, people do not look at assemblies critically enough,” she says.
Assessing assemblers
Right now, when researchers describe a new assembler, they often run it on a new data set, making comparisons difficult. But a few projects are examining how different assemblers perform with the same data. The goal is to learn both how to assemble high-quality genomes and how to recognize a high-quality assembly. For the Genome Assembly Gold-standard Evaluations (GAGE), scientists led by Steven Salzberg at Johns Hopkins University School of Medicine assembled four genomes (three of which had been previously published) using eight of the most popular de novo assembly algorithms3.
Two other efforts, the Assemblathon and dnGASP (de novo Genome Assembly Assessment Project), have taken the form of competitions. Teams generally consisted of the software designers for particular assemblers, who could adjust parameters as they thought best before submitting assembled genomes for evaluation. Performance was evaluated using simulated data from computer-designed genomes4.
The point is not identifying the best overall assembler at a particular point in time, but finding ways to assess and improve assemblers in general, says Ivo Gut, director of the National Genome Analysis Center in Spain, who ran dnGASP. dnGASP compared assembly teams' performance on a specially designed set of artificial chromosomes: three derived from the human genome, three from the chicken genome, and others representing the fruit fly, nematode, brewer's yeast and two species of mustard plant. In addition, the contest organizers included special 'challenge chromosomes' that tested assembler performance on various repetitive structures, divergent alleles and other difficult content.

The data set for these calibration chromosomes should be freely available later this year. “You can run [the reference data set] through your assembler and post the results back on the server. And then you can optimize your results,” says Gut. Researchers can tune assembly parameters for their genome of interest and benchmark the performance of new versions of their assemblers, getting an early indication of an assembler's performance with a modest investment of computational time, he explains. The calibration set is intended for assemblers that run on data produced by Illumina sequencers.

But optimization on artificial data may not be optimization at all, says Jaffe. “It's not a good use [of resources] to optimize on things that are different from reality,” he says. No matter how much effort is put into mimicking experimental artifacts and biases, simulated data won't mirror actual data well, he says. Salzberg agrees. “Some assemblers perform beautifully on simulated data but fall down on actual data,” he says.
Assemblathon organizer Korf admits that simulated data are not ideal, but says they offer a huge practical advantage. Even the best published reference genomes contain mistakes, and contests require the use of unpublished data, he says. “For judging assembly programs, it's nice [for the judges] to know the answer.” But Korf says Assemblathon 1 and the proliferation of sequencing projects have literally changed the game. Assemblathon 2 supplied real sequencing data for a parrot, cichlid fish and boa constrictor whose genomes have not yet been published. Results should be announced later this year.
The Assemblathon, dnGASP and GAGE all showed that even the best assemblers make numerous and important errors. Biologists working with genomes of newly sequenced species should remember that, says Salzberg. “All these assemblies are drafts. The quality is not anywhere close to what you were getting from Sanger assemblies.” Newer assemblies may be a hundredth to a thousandth the cost, he says, “but people are going to be very disappointed if they expect too much [from them].”
Benedict Paten, a computational biologist at the University of California, Santa Cruz, who is analyzing Assemblathon results in hopes of applying lessons to vertebrate sequencing projects, has a more optimistic view. “I was concerned when I started it that it was all hype,” he says, “but I was reassured, for the amount of money, by just how good the assemblies could be.”

But determining how good an assembly is can be difficult. “We would love to be able to do that,” says Korf, “to say 'That's a 5-star genome',” but reality is more complicated. The Assemblathon evaluated some hundred metrics in terms of how complete and accurate the assemblies were. Though N50 did correlate, roughly, with genome quality, scientists concluded that no set of metrics was perfect. “People have used these as a yardstick for quality, but that's naive,” says Paten. “There is no way to come up with a single best metric.”
Improvements in one metric often come at the expense of others. So-called conservative assemblers require extensive overlap and robust data to join reads into contigs and contigs into scaffolds; they have lower error rates, but the contigs and scaffolds are much shorter and so reveal less about how a genome is organized. Aggressive assemblers produce longer contigs and scaffolds but are more likely to join regions in the wrong order and orientation. Though researchers are working out ways to identify telltale signs of misassemblies and correct them, errors are hard to detect.
In addition to trying to find metrics or a set of metrics that serve as surrogates for assembly quality, researchers should see how accurately assemblers perform with data sets for well-characterized genomes, like mouse and human, says Jaffe. And other assessments need to be more standardized, he says: most assembly papers analyze how many genes are identified in a genome, or how much of a transcriptome can be found in an assembly, but methods of analysis vary so much that results are hard to interpret and hard to compare.
Organism adjustments
The accuracy and completeness of a genome assembly depends not just on computer programs and sequencing technologies, but also on the characteristics of the genome to be assembled. GAGE evaluated assemblers using data on two relatively complicated bacterial genomes (one harboring multiple plasmids), human chromosome 14 and the bumblebee genome. For human chromosome 14, one widely used assembler omitted only 1.7% of the reference assembly sequence. But although it placed first for this metric with the human chromosome, the program was in last place for Rhodobacter sphaeroides, omitting 7.5% of the genome.
The best approach to genome assembly varies by organism, says Daniel Rokhsar, eukaryotic group leader at the Joint Genome Institute, who has worked on plant, fungal and animal genomes. Solutions that are routine for small bacterial genomes are impossible or impractical for eukaryotes. Laboratory strains of worms, fish and mice are often so inbred that there is little heterozygosity, simplifying sequence assembly. And the size, spacing and arrangement of repetitive regions vary in ways that can trip up assemblers. “There are realities that come into play that can be genome dependent in ways that we don't really understand,” says Rokhsar.
The sea squirt is a good example. Human genomes vary between individuals at a rate of about 1 base pair per 1,000, explains Rokhsar. Sea squirts are much more polymorphic: about 1 in every 50–100 base pairs differ between homologous chromosomes. When an assembler encounters reads that are slightly different from each other, it must decide whether the reads are derived from the same locus or from repetitive regions. Faulty assumptions about rates of polymorphism can cause assemblers to drop genes, particularly members of closely related gene families.
Current data formats force genome assemblies to be inaccurate, says Jaffe. For example, he says, it may be clear that a locus contains a string of thymines, but not how many. Or scientists may know that a section is repeated but not how many times, or that one locus is highly variable. Such information is hard to include in an assembly because the current data format, called FASTA, cannot represent such uncertainties, explains Jaffe, who is working with the Assemblathon group on a new version, called FASTG, that will “let assemblers show what the possibilities are.”
Taking up the right data
GAGE evaluated how much better genome assemblers performed if protocols included additional error-correction steps to remove faulty reads before assembly. Sequencing machines themselves are equipped with computational filters to remove misread sequences as well as those that contain sequences from artifacts of library preparation and contaminating bacteria, but these filters aren't perfect, says Salzberg. “If you have a read that has even two or three bases that don't belong, then as soon as that read gets put into a contig, the contig can't be extended.” Even small errors on 1% of the relevant reads can break up the assembly, he says. Including additional error-correction algorithms produced bigger N50s and also reduced rates of misjoining.
And some of the most important work occurs before sequencing starts. Researchers should make sure to minimize sequencing errors, get high enough coverage, and get long enough reads and properly spaced paired reads. “Everyone is talking about the assembly, but that depends on the sequencing, and that depends on the library [which is fed into the sequencer],” says Korf.

Assemblies might also improve if assemblers were designed to incorporate more kinds of data. “We used transcribed RNA in the past to correct and improve Sanger assemblies,” says Salzberg, “but it is not yet part of any production-ready assembler [that uses short reads].” As part of Assemblathon 2, contestants had access to multiple kinds of data, such as sequence reads produced by different sequencing technologies. However, data outside short reads are underutilized, says Korf.
Dealing with ambiguity
Biologists can still use overall metrics to get a sense of how cautious to be, says Gene Robinson, an entomologist at the University of Illinois in Urbana-Champaign, who worked with Salzberg on GAGE and is also leading the i5K effort. “Two things that biologists need to know about de novo assembly are 'How much of the genome is estimated to be included in the assembly?' and 'How many different unconnected pieces does the assembly involve?'” he says. Those parameters indicate how easy it will be to perform comparative, functional and evolutionary analyses on a genome sequence.
If a particular area of the genome seems to be poorly assembled, targeted sequencing could be considered, says Felsenfeld. In some cases, it will make sense to home in on genomic regions of high biological interest, perhaps propagating certain regions in fosmids or bacterial artificial chromosomes. Such studies are too expensive to be conducted over the whole genome, he says, but could be worthwhile for some regions. “Perfection is impractical,” he says. “Do the best you can, and then refine it.”

No matter what sequencing technology is used to build a genome, biologists should try to anticipate just what might be wrong with their assemblies (Box 1). “Realistically speaking, most genomes are never going to get the polishing that human and mouse got,” says Church. And even those well-characterized genomes contain major omissions. 'Polishing' efforts on the final mouse genome revealed nearly 140 megabases of new sequence. Moreover, thousands of genes in the refined regions were evolutionarily recent and specific to mice; high levels of duplication had made them resistant to assembly5. In other work, Evan Eichler of the University of Washington and colleagues found that a de novo assembly of a human genome was missing 420 megabases of repeated sequence and over 2,000 protein-coding exons6.
Ideally, genomicists should work out more metrics for particular regions of the genome, not just for the genome as a whole, says Felsenfeld. “People like to talk about an absolute quality, but there is none,” he says. “You have to ask about the quality relative to likely uses.”
References
Pagani, I. et al. Nucleic Acids Res. 40, D571–579 (2012).
Parra, G., Bradnam, K., Ning, Z., Keane, T. & Korf, I. Nucleic Acids Res. 37, 289–297 (2009).
Salzberg, S.L. et al. Genome Res. 22, 557–567 (2012).
Earl, D. et al. Genome Res. 21, 2224–2241 (2011).
Church, D.M. et al. PLoS Biol. 7, e1000112 (2009).
Alkan, C., Sajjadian S. & Eichler, E.E. Nat. Methods 8, 61–65 (2011).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Baker, M. De novo genome assembly: what every biologist should know. Nat Methods 9, 333–337 (2012). https://doi.org/10.1038/nmeth.1935
Published:
Issue Date:
DOI: https://doi.org/10.1038/nmeth.1935
This article is cited by
-
Localized assembly for long reads enables genome-wide analysis of repetitive regions at single-base resolution in human genomes
Human Genomics (2023)
-
Identification and high-throughput genotyping of single nucleotide polymorphism markers in a non-model conifer (Abies nordmanniana (Steven) Spach)
Scientific Reports (2023)
-
Toward a more holistic method of genome assembly assessment
BMC Bioinformatics (2020)
-
Raw transcriptomics data to gene specific SSRs: a validated free bioinformatics workflow for biologists
Scientific Reports (2020)
-
Tools for Chrysanthemum genetic research and breeding: Is genotyping-by-sequencing (GBS) the best approach?
Horticulture, Environment, and Biotechnology (2019)
