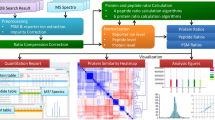
Abstract
We here describe a normalization method to combine quantitative proteomics data. By merging the output of two popular quantification software packages, we obtained a 20% increase (on average) in the number of quantified human proteins without suffering from a loss of quality. Our integrative workflow is freely available through our user-friendly, open-source Rover software (http://compomics-rover.googlecode.com/).
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Domon, B. & Aebersold, R. Science 312, 212–217 (2006).
Vaudel, M., Sickmann, A. & Martens, L. Proteomics 10, 650–670 (2010).
Mueller, L.N., Brusniak, M., Mani, D.R. & Aebersold, R. J. Proteome Res. 7, 51–61 (2008).
Cox, J. & Mann, M. Nat. Biotechnol. 26, 1367–1372 (2008).
Mortensen, P. et al. J. Proteome Res. 9, 393–403 (2010).
Park, S.K., Venable, J.D., Xu, T. & Yates, J.R. III. Nat. Methods 5, 319–322 (2008).
Colaert, N., Vandekerckhove, J., Martens, L. & Gevaert, K. in Methods in Molecular Biology: Gel-Free Proteomics: Methods and Protocols (eds. Gevaert, K. and Vandekerckhove, J.) (Humana Press; in the press).
Yu, W. et al. Proteomics 10, 1172–1189 (2010).
Jones, A.R., Siepen, J.A., Hubbard, S.J. & Paton, N.W. Proteomics 9, 1220–1229 (2009).
Searle, B.C., Turner, M. & Nesvizhskii, A.I. J. Proteome Res. 7, 245–253 (2008).
Ong, S.-E. et al. Mol. Cell. Proteomics 1, 376–386 (2002).
Gevaert, K. et al. Mol. Cell. Proteomics 1, 896–903 (2002).
Bartke, T. et al. Cell 143, 470–484 (2010).
Yang, Y.H. et al. Nucleic Acids Res. 30, E15 (2002).
Colaert, N., Helsens, K., Impens, F., Vandekerckhove, J. & Gevaert, K. Proteomics 10, 1226–1229 (2010).
Perkins, D.N., Pappin, D.J., Creasy, D.M. & Cottrell, J.S. Electrophoresis 20, 3551–3567 (1999).
Acknowledgements
C.V.H. is supported by a grant of the Research Foundation–Flanders (project 3G003908). We thank B. Ghesquière, F. Impens and E. Timmerman for providing prepublication access during algorithm development to their now published data. We acknowledge the support of Ghent University (Multidisciplinary Research Partnership “Bioinformatics: from nucleotides to networks”) and EU 7th Framework Programme (contract 262067-PRIME-XS). K.G. and J.V. acknowledge funding from the Fund for Scientific Research–Flanders (Belgium) (project G.0042.07), the Concerted Research Actions (project BOF07/GOA/012) from Ghent University and the Interuniversity Attraction Poles (IUAP06).
Author information
Authors and Affiliations
Contributions
N.C. developed the combination algorithm and wrote the first draft of the manuscript. C.V.H. contributed to algorithm development and to manuscript writing. S.D. contributed to algorithm development and manuscript writing. A.S. assisted with data processing and manuscript writing. J.V. and K.G. supervised part of the work and contributed to manuscript writing. L.M. supervised the work, contributed to algorithm development and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–3, Supplementary Table 1–2, Supplementary Data (PDF 11502 kb)
Rights and permissions
About this article
Cite this article
Colaert, N., Van Huele, C., Degroeve, S. et al. Combining quantitative proteomics data processing workflows for greater sensitivity. Nat Methods 8, 481–483 (2011). https://doi.org/10.1038/nmeth.1604
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nmeth.1604
This article is cited by
-
Towards a human proteomics atlas
Analytical and Bioanalytical Chemistry (2012)