Abstract
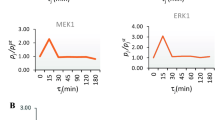
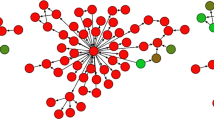
We describe microwestern arrays, which enable quantitative, sensitive and high-throughput assessment of protein abundance and modifications after electrophoretic separation of microarrayed cell lysates. This method allowed us to measure 91 phosphosites on 67 proteins at six time points after stimulation with five epidermal growth factor (EGF) concentrations in A431 human carcinoma cells. We inferred the connectivities among 15 phosphorylation sites in 10 receptor tyrosine kinases (RTKs) and two sites from Src kinase using Bayesian network modeling and two mutual information-based methods; the three inference methods yielded substantial agreement on the network topology. These results imply multiple distinct RTK coactivation mechanisms and support the notion that small amounts of experimental data collected from phenotypically diverse network states may enable network inference.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Burnette, W. “Western blotting”: electrophoretic transfer of proteins from sodium dodecyl sulfate polyacrylamide gels to unmodified nitrocellulose and radiographic detection with antibody and radioiodinated protein A. Anal. Biochem. 112, 195–203 (1981).
Paweletz, C.P., Liotta, L.A. & Petricoin, E.F. New technologies for biomarker analysis of prostate cancer progression: Laser capture microdissection and tissue proteomics. Urology 57, 160–163 (2001).
Paweletz, C.P. et al. Reverse phase protein microarrays which capture disease progression show activation of pro-survival pathways at the cancer invasion front. Oncogene 20, 1981–1989 (2001).
Sevecka, M. & MacBeath, G. State-based discovery: a multidimensional screen for small-molecule modulators of EGF signaling. Nat. Methods 3, 825–831 (2006).
Sachs, K., Perez, O., Pe′er, D., Lauffenburger, D.A. & Nolan, G.P. Causal protein-signaling networks derived from multiparameter single-cell data. Science 308, 523–529 (2005).
Rikova, K. et al. Global survey of phosphotyrosine signaling identifies oncogenic kinases in lung cancer. Cell 131, 1190–1203 (2007).
Olsen, J.V. et al. Global, in vivo and site-specific phosphorylation dynamics in signaling networks. Cell 127, 635–648 (2006).
Wolf-Yadlin, A. et al. Effects of HER2 overexpression on cell signaling networks governing proliferation and migration. Mol. Syst. Biol. 2, 54 (2006).
Tibes, R. et al. Reverse phase protein array: validation of a novel proteomic technology and utility for analysis of primary leukemia specimens and hematopoietic stem cells. Mol. Cancer Ther. 5, 2512–2521 (2006).
Jones, R.B., Gordus, A., Krall, J.A. & MacBeath, G. A quantitative protein interaction network for the ErbB receptors using protein microarrays. Nature 439, 168–174 (2006).
Sunada, H., Magun, B.E., Mendelsohn, J. & MacLeod, C.L. Monoclonal antibody against epidermal growth factor receptor is internalized without stimulating receptor phosphorylation. Proc. Natl. Acad. Sci. USA 83, 3825–3829 (1986).
Gill, G.N. & Lazar, C.S. Increased phosphotyrosine content and inhibition of proliferation in EGF-treated A431 cells. Nature 293, 305–307 (1981).
Wolf-Yadlin, A., Hautaniemi, S., Lauffenburger, D.A. & White, F.M. Multiple reaction monitoring for robust quantitative proteomic analysis of cellular signaling networks. Proc. Natl. Acad. Sci. USA 104, 5860–5865 (2007).
Chang, F. et al. Involvement of PI3K//Akt pathway in cell cycle progression, apoptosis and neoplastic transformation: a target for cancer chemotherapy. Leukemia 17, 590–603 (2003).
Pe'er, D. Bayesian network analysis of signaling networks: a primer. Sci. STKE 2005, l4 (2005).
Koivisto, M. & Sood, K. Exact Bayesian structure discovery in Bayesian networks. J. Mach. Learn. Res. 5, 549–573 (2004).
Eaton, D. & Murphy, K. Exact Bayesian structure learning from uncertain interventions. in Proc. 12th Conf. on AI and Stats, 107–114 (2007).
Chickering, D.M. Learning equivalence classes of bayesian-network structures. J. Mach. Learn. Res. 2, 445–498 (2002).
Downward, J., Parker, P. & Waterfield, M.D. Autophosphorylation sites on the epidermal growth factor receptor. Nature 311, 483–485 (1984).
Saito, Y., Haendeler, J., Hojo, Y., Yamamoto, K. & Berk, B.C. Receptor heterodimerization: essential mechanism for platelet-derived growth factor-induced epidermal growth factor receptor transactivation. Mol. Cell. Biol. 21, 6387–6394 (2001).
Duchesne, L., Tissot, B., Rudd, T.R., Dell, A. & Fernig, D.G. N-glycosylation of fibroblast growth factor receptor 1 regulates ligand and heparan sulfate co-receptor binding. J. Biol. Chem. 281, 27178–27189 (2006).
Ekman, S., Kallin, A., Engström, U., Heldin, C. & Rönnstrand, L. SHP-2 is involved in heterodimer specific loss of phosphorylation of Tyr771 in the PDGF beta-receptor. Oncogene 21, 1870–1875 (2002).
Gould, K.L. & Hunter, T. Platelet-derived growth factor induces multisite phosphorylation of pp60c-src and increases its protein-tyrosine kinase activity. Mol. Cell. Biol. 8, 3345–3356 (1988).
Margolin, A.A. et al. ARACNE: an algorithm for the reconstruction of gene regulatory networks in a mammalian cellular context. BMC Bioinformatics 7 (Suppl. 1), S7 (2006).
Faith, J.J. et al. Large-scale mapping and validation of Escherichia coli transcriptional regulation from a compendium of expression profiles. PLoS Biol. 5, e8 (2007).
Taylor, R.C., Acquaah-Mensah, G., Singhal, M., Malhotra, D. & Biswal, S. Network inference algorithms elucidate Nrf2 regulation of mouse lung oxidative stress. PLOS Comput. Biol. 4, e1000166 (2008).
Cantone, I. et al. A yeast synthetic network for in vivo assessment of reverse-engineering and modeling approaches. Cell 137, 172–181 (2009).
Husmeier, D. Sensitivity and specificity of inferring genetic regulatory interactions from microarray experiments with dynamic Bayesian networks. Bioinformatics 19, 2271–2282 (2003).
Lou, Y.W. et al. Redox regulation of the protein tyrosine phosphatase PTP1B in cancer cells. FEBS J. 275, 69–88 (2008).
Lu, W., Shen, K. & Cole, P.A. Chemical dissection of the effects of tyrosine phosphorylation of SHP-2. Biochemistry 42, 5461–5468 (2003).
Friedman, N. & Koller, D. Being Bayesian about Bayesian network structure: a Bayesian approach to structure discovery in Bayesian networks. Mach. Learn. 50, 95–125 (2003).
Heckerman, D., Geiger, D. & Chickering, D.M. Learning Bayesian networks: The combination of knowledge and statistical data. Mach. Learn. 20, 197–243 (1995).
Madigan, D. & York, J. Bayesian graphical models for discrete data. Int. Stat. Rev. 63, 215–232 (1995).
Meyer, P., Lafitte, F. & Bontempi, G. minet: a R/Bioconductor package for inferring large transcriptional networks using mutual information. BMC Bioinformatics 9, 461 (2008).
Acknowledgements
A431 cells were kindly provided by S. Liao (The University of Chicago). We thank C.Y. Chuang for technical assistance with MWAs, M.R. Rosner, K.P. White and W.L. McKeehan for helpful discussions, C. May for the graphic design of Figure 1, and J. Barkinge for operating support with microarraying. This work was supported, in part, by awards from The University of Chicago Cancer Research Center, The American Cancer Society, The University of Chicago Breast Cancer Specialized Program of Research Excellence, The Cancer Research Foundation and The Illinois Department of Health (86280156 to R.B.J.), the US National Institutes of General Medical Sciences P50-GM0686762 Cell Decision Processes Center grant and US National Cancer Institute CA96504 to D.A.L.; M.F.C. was supported by a National Institutes of Health Systems Biology of Oxygen Predoctoral Training grant; J.P.W. was supported by a National Science Foundation Graduate fellowship; and C.-P.C. was supported by a National Institutes of Health Cancer Biology Postdoctoral Training grant.
Author information
Authors and Affiliations
Contributions
C.-P.C., M.F.C. and R.B.J. designed the experiments. C.-P.C. and M.F.C. performed the cell culture and growth factor stimulations. M.F.C. and R.B.J. designed the MWA method, M.F.C. carried out microwestern experiments and organized the data into heat maps. J.P.W. and D.A.L. performed Bayesian network, CLR and ARACNe analysis of the data. M.F.C., J.P.W., D.A.L. and R.B.J. wrote the manuscript. All authors read and revised the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–12, Supplementary Notes 1–3, Supplementary Tables 3,5 (PDF 4837 kb)
Supplementary Table 1
Fold changes, standard deviations, signal/background ratios, antibody catalog numbers, protein sizes and local epitope sequence for protein abundances and modifications quantified by microwestern arrays. (XLS 285 kb)
Supplementary Table 2
Antibody layout for Figure 4. (XLS 32 kb)
Supplementary Table 4
The epitope recognition sequence surrounding the 20 phosphosites for which signals were quantified in this study compared to the sequences of all 57 human-encoded receptor tyrosine kinases. (XLS 37 kb)
Rights and permissions
About this article
Cite this article
Ciaccio, M., Wagner, J., Chuu, CP. et al. Systems analysis of EGF receptor signaling dynamics with microwestern arrays. Nat Methods 7, 148–155 (2010). https://doi.org/10.1038/nmeth.1418
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nmeth.1418
This article is cited by
-
Practical application of a Bayesian network approach to poultry epigenetics and stress
BMC Bioinformatics (2022)
-
Screening of organoids derived from patients with breast cancer implicates the repressor NCOR2 in cytotoxic stress response and antitumor immunity
Nature Cancer (2022)
-
ROR2 suppresses metastasis of prostate cancer via regulation of miR-199a-5p–PIAS3–AKT2 signaling axis
Cell Death & Disease (2020)
-
Reconstructing kinase network topologies from phosphoproteomics data reveals cancer-associated rewiring
Nature Biotechnology (2020)
-
Tracking the expression of therapeutic protein targets in rare cells by antibody-mediated nanoparticle labelling and magnetic sorting
Nature Biomedical Engineering (2020)