Abstract
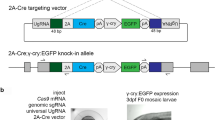
We describe a method for the highly efficient and precise targeted modification of gene trap loci in mouse embryonic stem cells (ESCs). Through the Floxin method, gene trap mutations were reverted and new DNA sequences inserted using Cre recombinase and a shuttle vector, pFloxin. Floxin technology is applicable to the existing collection of 24,149 compatible gene trap cell lines, which should enable high-throughput modification of many genes in mouse ESCs.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Stanford, W.L., Epp, T., Reid, T. & Rossant, J. Methods Enzymol. 420, 136–162 (2006).
Hansen, J. et al. Proc. Natl. Acad. Sci. USA 100, 9918–9922 (2003).
Araki, K. et al. Cell. Mol. Biol. (Noisy-le-grand, France) 45, 737–750 (1999).
Hardouin, N. & Nagy, A. Genesis 26, 245–252 (2000).
Osipovich, A.B., Singh, A. & Ruley, H.E. Genome Res. 15, 428–435 (2005).
Schnutgen, F. et al. Proc. Natl. Acad. Sci. USA 102, 7221–7226 (2005).
Taniwaki, T. et al. Dev. Growth Differ. 47, 163–172 (2005).
Xin, H.B. et al. Nucleic Acids Res. 33, e14 (2005).
Sauer, B. Methods Enzymol. 225, 890–900 (1993).
Albert, H., Dale, E.C., Lee, E. & Ow, D.W. Plant J. 7, 649–659 (1995).
Skarnes, W.C. et al. Nat. Genet. 36, 543–544 (2004).
Ferrante, M.I. et al. Nat. Genet. 38, 112–117 (2006).
Pasini, D., Bracken, A.P., Hansen, J.B., Capillo, M. & Helin, K. Mol. Cell. Biol. 27, 3769–3779 (2007).
Raymond, C.S. & Soriano, P. PLoS One 2, e162 (2007).
Nagy, A., Rossant, J., Nagy, R., Abramow-Newerly, W. & Roder, J.C. Proc. Natl. Acad. Sci. USA 90, 8424–8428 (1993).
Gu, H., Zou, Y.R. & Rajewsky, K. Cell 73, 1155–1164 (1993).
Acknowledgements
We thank K. Thorn for assistance with confocal microscopy, and the members of the Reiter lab for critical reading. Grants from the US National Science Foundation (V.S. and N.S.), US National Institutes of Health (RO1AR054396), California Institute for Regenerative Medicine (RN2-00919), the Burroughs Wellcome Fund, the Packard Foundation, the Leona M. and Harry B. Helmsley Charitable Trust, and the Sandler Family Supporting Foundation (J.F.R.) funded this work. Suz12 and Sall4 cDNAs were gifts from M. Ramalho-Santos (University of California, San Francisco).
Author information
Authors and Affiliations
Contributions
V.S., J.H., W.C.S. and J.F.R. conceived and designed the experiments. P.W. conceived and designed the first gene trap–Floxin vector. V.S., J.H., N.S., A.D.S., W.C.S., A.R.N. and J.F.R. performed the experiments. J.F.R. and V.S. wrote the paper.
Corresponding authors
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–4 and Supplementary Tables 2–6 (PDF 3879 kb)
Supplementary Table 1
Floxin-compatible cell lines and corresponding trapped genes. (XLS 683 kb)
Rights and permissions
About this article
Cite this article
Singla, V., Hunkapiller, J., Santos, N. et al. Floxin, a resource for genetically engineering mouse ESCs. Nat Methods 7, 50–52 (2010). https://doi.org/10.1038/nmeth.1406
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nmeth.1406
This article is cited by
-
Wapl repression by Pax5 promotes V gene recombination by Igh loop extrusion
Nature (2020)
-
Knockin of Cre Gene at Ins2 Locus Reveals No Cre Activity in Mouse Hypothalamic Neurons
Scientific Reports (2016)
-
Dual RMCE for efficient re-engineering of mouse mutant alleles
Nature Methods (2010)