Abstract
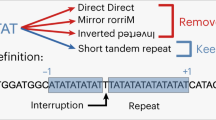
Metazoan genomes contain thousands of sequence tracts that match the guanine-quadruplex (G4) DNA signature G3NxG3NxG3NxG3, a motif that is intrinsically mutagenic, probably because it can form secondary structures during DNA replication. Here we show how and to what extent this feature can be used to generate deletion alleles of many Caenorhabditis elegans genes.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Maizels, N. Nat. Struct. Mol. Biol. 13, 1055–1059 (2006).
Voineagu, I., Narayanan, V., Lobachev, K.S. & Mirkin, S.M. Proc. Natl. Acad. Sci. USA 105, 9936–9941 (2008).
Tornaletti, S., Park-Snyder, S. & Hanawalt, P.C. J. Biol. Chem. 283, 12756–12762 (2008).
Gellert, M., Lipsett, M.N. & Davies, D.R. Proc. Natl. Acad. Sci. USA 48, 2013–2018 (1962).
Sen, D. & Gilbert, W. Nature 334, 364–366 (1988).
Cheung, I. et al. Nat. Genet. 31, 405–409 (2002).
Youds, J.L. et al. Mol. Cell. Biol. 28, 1470–1479 (2008).
Kruisselbrink, E. et al. Curr. Biol. 18, 900–905 (2008).
Edgley, M. et al. Nucleic Acids Res. 30, e52 (2002).
Gengyo-Ando, K. & Mitani, S. Biochem. Biophys. Res. Commun. 269, 64–69 (2000).
Acknowledgements
We thank members of the Tijsterman lab for discussions, R. Plasterk for generous support and WormBase curators for building the platform that allows C. elegans genomic research. This work was funded by a Horizon grant from the Netherlands Genomics Initiative and a Vidi grant from ZonMw to M.T.
Author information
Authors and Affiliations
Contributions
D.B.P., E.K. and M.T. performed the experiments and analyzed the data. V.G. performed the bioinformatics analysis. M.T. conceived and supervised the project, and wrote the paper.
Corresponding author
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–2 and Supplementary Table 1 (PDF 528 kb)
Supplementary Table 2
C. elegans endogenous G4- sites (XLS 475 kb)
Rights and permissions
About this article
Cite this article
Pontier, D., Kruisselbrink, E., Guryev, V. et al. Isolation of deletion alleles by G4 DNA-induced mutagenesis. Nat Methods 6, 655–657 (2009). https://doi.org/10.1038/nmeth.1362
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nmeth.1362
This article is cited by
-
Mechanical insights into ribosomal progression overcoming RNA G-quadruplex from periodical translation suppression in cells
Scientific Reports (2016)
-
Spectrum of variations in dog-1/FANCJ and mdf-1/MAD1 defective Caenorhabditis elegans strains after long-term propagation
BMC Genomics (2015)
-
Mutagenic consequences of a single G-quadruplex demonstrate mitotic inheritance of DNA replication fork barriers
Nature Communications (2015)
-
A Polymerase Theta-dependent repair pathway suppresses extensive genomic instability at endogenous G4 DNA sites
Nature Communications (2014)
-
DNA secondary structures and epigenetic determinants of cancer genome evolution
Nature Structural & Molecular Biology (2011)