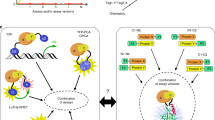
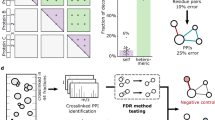
Abstract
Information on protein-protein interactions is of central importance for many areas of biomedical research. At present no method exists to systematically and experimentally assess the quality of individual interactions reported in interaction mapping experiments. To provide a standardized confidence-scoring method that can be applied to tens of thousands of protein interactions, we have developed an interaction tool kit consisting of four complementary, high-throughput protein interaction assays. We benchmarked these assays against positive and random reference sets consisting of well documented pairs of interacting human proteins and randomly chosen protein pairs, respectively. A logistic regression model was trained using the data from these reference sets to combine the assay outputs and calculate the probability that any newly identified interaction pair is a true biophysical interaction once it has been tested in the tool kit. This general approach will allow a systematic and empirical assignment of confidence scores to all individual protein-protein interactions in interactome networks.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Barrios-Rodiles, M. et al. High-throughput mapping of a dynamic signaling network in mammalian cells. Science 307, 1621–1625 (2005).
von Mering, C. et al. Comparative assessment of large-scale data sets of protein-protein interactions. Nature 417, 399–403 (2002).
Yu, H. et al. High-quality binary protein interaction map of the yeast interactome network. Science 322, 104–110 (2008).
Ge, H., Liu, Z., Church, G.M. & Vidal, M. Correlation between transcriptome and interactome mapping data from Saccharomyces cerevisiae. Nat. Genet. 29, 482–486 (2001).
Ramani, A.K., Bunescu, R.C., Mooney, R.J. & Marcotte, E.M. Consolidating the set of known human protein-protein interactions in preparation for large-scale mapping of the human interactome. Genome Biol. 6, R40 (2005).
Ramani, A.K. et al. A map of human protein interactions derived from co-expression of human mRNAs and their orthologs. Mol. Syst. Biol. 4, 180 (2008).
Chiang, T. et al. Coverage and error models of protein-protein interaction data by directed graph analysis. Genome Biol. 8, R186 (2007).
Ito, T. et al. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc. Natl. Acad. Sci. USA 98, 4569–4574 (2001).
Sato, S. et al. A large-scale protein protein interaction analysis in Synechocystis sp. PCC6803. DNA Res. 14, 207–216 (2007).
Gavin, A.C. et al. Proteome survey reveals modularity of the yeast cell machinery. Nature 440, 631–636 (2006).
Krogan, N.J. et al. Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature 440, 637–643 (2006).
Han, J.D. et al. Evidence for dynamically organized modularity in the yeast protein-protein interaction network. Nature 430, 88–93 (2004).
Li, S. et al. A map of the interactome network of the metazoan C. elegans. Science 303, 540–543 (2004).
Rual, J.F. et al. Towards a proteome-scale map of the human protein-protein interaction network. Nature 437, 1173–1178 (2005).
Vidalain, P.O. et al. Increasing specificity in high-throughput yeast two-hybrid experiments. Methods 32, 363–370 (2004).
Walhout, A.J. & Vidal, M. High-throughput yeast two-hybrid assays for large-scale protein interaction mapping. Methods 24, 297–306 (2001).
Eyckerman, S. et al. Design and application of a cytokine-receptor-based interaction trap. Nat. Cell Biol. 3, 1114–1119 (2001).
Nyfeler, B., Michnick, S.W. & Hauri, H.P. Capturing protein interactions in the secretory pathway of living cells. Proc. Natl. Acad. Sci. USA 102, 6350–6355 (2005).
Ramachandran, N. et al. Next-generation high-density self-assembling functional protein arrays. Nat. Methods 5, 535–538 (2008).
Venkatesan, K. et al. An empirical framework for binary interactome mapping. Nat. Methods advance online publication, 10.1038/nmeth.1280 (7 December 2008).
Bader, G.D., Betel, D. & Hogue, C.W. BIND: the Biomolecular Interaction Network Database. Nucleic Acids Res. 31, 248–250 (2003).
Chatr-aryamontri, A. et al. MINT: the Molecular INTeraction database. Nucleic Acids Res. 35, D572–D574 (2007).
Mishra, G.R. et al. Human protein reference database–2006 update. Nucleic Acids Res. 34, D411–D414 (2006).
Pagel, P. et al. The MIPS mammalian protein-protein interaction database. Bioinformatics 21, 832–834 (2005).
Salwinski, L. et al. The Database of Interacting Proteins: 2004 update. Nucleic Acids Res. 32, D449–D451 (2004).
Cusick, M.E. et al. Literature-curated protein interaction datasets. Nat. Methods (in the press).
Rual, J.F. et al. Human ORFeome version 1.1: a platform for reverse proteomics. Genome Res. 14, 2128–2135 (2004).
Ben-Hur, A. & Noble, W.S. Choosing negative examples for the prediction of protein-protein interactions. BMC Bioinformatics 7 (suppl. 1), S2 (2006).
Qi, Y., Bar-Joseph, Z. & Klein-Seetharaman, J. Evaluation of different biological data and computational classification methods for use in protein interaction prediction. Proteins 63, 490–500 (2006).
Vidal, M. et al. Reverse two-hybrid and one-hybrid systems to detect dissociation of protein-protein and DNA-protein interactions. Proc. Natl. Acad. Sci. USA 93, 10315–10320 (1996).
Lemmens, I. et al. Heteromeric MAPPIT: a novel strategy to study modification-dependent protein-protein interactions in mammalian cells. Nucleic Acids Res. 31, e75 (2003).
Lemmens, I., Lievens, S., Eyckerman, S. & Tavernier, J. Reverse MAPPIT detects disruptors of protein-protein interactions in human cells. Nat. Protoc. 1, 92–97 (2006).
Acknowledgements
We thank S. Michnick and N. Ramachandran for reagents and technical help for the PCA and wNAPPA assays, respectively. We thank A. Datti, T. Sun and F. Vizeacoumar from the SMART Robotics Facility at the Samuel Lunenfeld Research Institute for help with the automated version of LUMIER assay. We thank all members of the Vidal, Tavernier, Roth, and Wrana laboratories for helpful discussions, Agencourt Biosciences for sequencing assistance, and A. Bird and D. Maher for administrative assistance. This work was supported by contributions from the W.M. Keck Foundation awarded to M.V., F.P.R. and D.E.H.; by the Ellison Foundation awarded to M.V.; by Institute Sponsored Research funds from the Dana-Farber Cancer Institute Strategic Initiative awarded to M.V. and CCSB; by US National Institutes of Health grants 5P50HG004233 and 2R01HG001715 awarded to M.V., F.P.R. and D.E.H., R01 ES015728 awarded to M.V., 5U54CA112952 awarded to J. Nevins (M.V. subcontract), 5U01CA105423 awarded to S.H. Orkin (M.V. project), R01 HG003224 awarded to F.P.R. and F32 HG004098 awarded to M.T.; by a University of Ghent grant GOA12051401 and the Fonds Wetenschappelijk Onderzoek– Vlanderen (FWO-V) G.0031.06 awarded to J.T., by a postdoctoral fellowship from the FWO-V awarded to I.L.; and by a grant from Genome Canada and funds from the Ontario Genomics Institute awarded to J.L.W. M.V. is a Chercheur Qualifié Honoraire from the Fonds de la Recherche Scientifique (FRS-FNRS, French Community of Belgium).
Author information
Authors and Affiliations
Contributions
P.B., M.T. and M.D. coordinated experiments and data analysis. P.B., M.D., J.M.S., J.-F.R., R.R.M. and H.Y. performed high-throughput Gateway cloning. P.B., H.Y. and J.M.S. implemented, developed and analyzed wNAPPA and PCA experiments. J.-F.R., K.V. and M.E.C. established PRSv1.0 and RRS reference sets. I.L., A.-S. de S., J.T. and K.V. coordinated, performed and analyzed MAPPIT experiments. M.B.-R., L.R. and J.L.W. coordinated, performed and analyzed LUMIER experiments. M.T. and F.P.R. developed the regression model. M.V. conceived the project. M.V., T.P., J.L.W. and D.E.H. developed the concepts underlying the overall strategy. D.E.H., F.P.R. and M.V. co-directed the project.
Corresponding authors
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–6, Supplementary Table 1, Supplementary Data, Supplementary Methods, Supplementary Protocols 1–5 (PDF 2582 kb)
Supplementary Table 2
PM_IDs and full names for all hsPRS_v1 protein pairs (XLS 77 kb)
Rights and permissions
About this article
Cite this article
Braun, P., Tasan, M., Dreze, M. et al. An experimentally derived confidence score for binary protein-protein interactions. Nat Methods 6, 91–97 (2009). https://doi.org/10.1038/nmeth.1281
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nmeth.1281
This article is cited by
-
A binary interaction map between turnip mosaic virus and Arabidopsis thaliana proteomes
Communications Biology (2023)
-
The protein interactome of the citrus Huanglongbing pathogen Candidatus Liberibacter asiaticus
Nature Communications (2023)
-
TIR Domains in Arabidopsis thaliana Suppressor of npr1-1, Constitutive 1 and Its Closely Related Disease Resistance Proteins Form Intricate Interaction Networks
Journal of Plant Biology (2023)
-
Next-generation large-scale binary protein interaction network for Drosophila melanogaster
Nature Communications (2023)
-
A proteome-scale map of the SARS-CoV-2–human contactome
Nature Biotechnology (2023)