Abstract
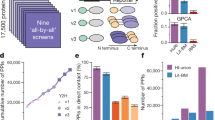
Several attempts have been made to systematically map protein-protein interaction, or 'interactome', networks. However, it remains difficult to assess the quality and coverage of existing data sets. Here we describe a framework that uses an empirically-based approach to rigorously dissect quality parameters of currently available human interactome maps. Our results indicate that high-throughput yeast two-hybrid (HT-Y2H) interactions for human proteins are more precise than literature-curated interactions supported by a single publication, suggesting that HT-Y2H is suitable to map a significant portion of the human interactome. We estimate that the human interactome contains ∼130,000 binary interactions, most of which remain to be mapped. Similar to estimates of DNA sequence data quality and genome size early in the Human Genome Project, estimates of protein interaction data quality and interactome size are crucial to establish the magnitude of the task of comprehensive human interactome mapping and to elucidate a path toward this goal.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Vidal, M. Interactome modeling. FEBS Lett. 579, 1834–1838 (2005).
Rual, J.F. et al. Towards a proteome-scale map of the human protein-protein interaction network. Nature 437, 1173–1178 (2005).
Stelzl, U. et al. A human protein-protein interaction network: a resource for annotating the proteome. Cell 122, 957–968 (2005).
Ewing, R.M. et al. Large-scale mapping of human protein-protein interactions by mass spectrometry. Mol. Syst. Biol. 3, 89 (2007).
Peri, S. et al. Human protein reference database as a discovery resource for proteomics. Nucleic Acids Res. 32, D497–D501 (2004).
Zanzoni, A. et al. MINT: a Molecular INTeraction database. FEBS Lett. 513, 135–140 (2002).
Bader, G.D. et al. BIND–The Biomolecular Interaction Network Database. Nucleic Acids Res. 29, 242–245 (2001).
Hermjakob, H. et al. IntAct: an open source molecular interaction database. Nucleic Acids Res. 32, D452–D455 (2004).
Xenarios, I. et al. DIP, the Database of Interacting Proteins: a research tool for studying cellular networks of protein interactions. Nucleic Acids Res. 30, 303–305 (2002).
Mewes, H.W. et al. MIPS: a database for genomes and protein sequences. Nucleic Acids Res. 30, 31–34 (2002).
Ramani, A.K., Bunescu, R.C., Mooney, R.J. & Marcotte, E.M. Consolidating the set of known human protein-protein interactions in preparation for large-scale mapping of the human interactome. Genome Biol. 6, R40 (2005).
Lehner, B. & Fraser, A.G. A first-draft human protein-interaction map. Genome Biol. 5, R63 (2004).
Hart, G.T., Ramani, A.K. & Marcotte, E.M. How complete are current yeast and human protein-interaction networks? Genome Biol. 7, 120 (2006).
Futschik, M.E., Chaurasia, G. & Herzel, H. Comparison of human protein-protein interaction maps. Bioinformatics 23, 605–611 (2007).
von Mering, C. et al. Comparative assessment of large-scale data sets of protein-protein interactions. Nature 417, 399–403 (2002).
Reguly, T. et al. Comprehensive curation and analysis of global interaction networks in Saccharomyces cerevisiae. J. Biol. 5, 11 (2006).
Gandhi, T.K. et al. Analysis of the human protein interactome and comparison with yeast, worm and fly interaction datasets. Nat. Genet. 38, 285–293 (2006).
Patil, A. & Nakamura, H. Filtering high-throughput protein-protein interaction data using a combination of genomic features. BMC Bioinformatics 6, 100 (2005).
Huang, H., Jedynak, B.M. & Bader, J.S. Where have all the interactions gone? Estimating the coverage of two-hybrid protein interaction maps. PLoS Comput. Biol. 3, e214 (2007).
D'Haeseleer, P. & Church, G.M. Estimating and improving protein interaction error rates. Proc. IEEE Comput. Syst. Bioinform. Conf. 216–223 (2004).
Grigoriev, A. On the number of protein-protein interactions in the yeast proteome. Nucleic Acids Res. 31, 4157–4161 (2003).
Deane, C.M., Salwinski, L., Xenarios, I. & Eisenberg, D. Protein interactions: two methods for assessment of the reliability of high throughput observations. Mol. Cell. Proteomics 1, 349–356 (2002).
Sprinzak, E., Sattath, S. & Margalit, H. How reliable are experimental protein-protein interaction data? J. Mol. Biol. 327, 919–923 (2003).
Rual, J.F. et al. Human ORFeome version 1.1: a platform for reverse proteomics. Genome Res. 14, 2128–2135 (2004).
Cusick, M.E. et al. Literature-curated protein interaction datasets. Nat. Methods (in the press).
Braun, P. et al. An experimentally derived confidence score for binary protein-protein interactions. Nat. Methods advance online publication, doi:10.1038/nmeth.1281 (7 December 2008).
Eyckerman, S. et al. Design and application of a cytokine-receptor-based interaction trap. Nat. Cell Biol. 3, 1114–1119 (2001).
Stumpf, M.P. et al. Estimating the size of the human interactome. Proc. Natl. Acad. Sci. USA 105, 6959–6964 (2008).
Ramírez, F., Schlicker, A., Assenov, Y., Lengauer, T. & Albrecht, M. Computational analysis of human protein interaction networks. Proteomics 7, 2541–2552 (2007).
Collins, S.R. et al. Towards a comprehensive atlas of the physical interactome of Saccharomyces cerevisiae. Mol. Cell. Proteomics 6, 439–450 (2007).
Fields, C., Adams, M.D., White, O. & Venter, J.C. How many genes in the human genome? Nat. Genet. 7, 345–346 (1994).
Lemmens, I., Lievens, S., Eyckerman, S. & Tavernier, J. Reverse MAPPIT detects disruptors of protein-protein interactions in human cells. Nat. Protoc. 1, 92–97 (2006).
Acknowledgements
We thank members of CCSB and the Vidal, Barabasi, Wanker and Tavernier laboratories and S. Sahasrabuddhe, R. Bell, R. Chettier and C. Wiggins for helpful discussions; E. Smith for help generating Figure 1; and Agencourt Biosciences for sequencing assistance. This work was supported by the US National Human Genome Research Institute (2R01HG001715 and 5P50HG004233 to M.V. and F.P.R.), the US National Cancer Institute (5U54CA112952 to J. Nevins, subcontract to M.V.; and 5U01CA105423 to S.H. Orkin, project to M.V.), the US National Institutes of Health (IH U01 A1070499-01 and U56 CA113004 to A.-L.B. and postdoctoral training grant fellowship T32CA09361 to K.V.), the Ellison Foundation (to M.V.), the W.M. Keck Foundation (to M.V.), Dana-Farber Cancer Institute Institute Sponsored Research funds (to M.V.), the US National Science Foundation (ITR DMR-0926737 and IIS-0513650 to A.-L.B.), Deutsches Bundesministerium für Bildung und Forschung (NGFN2, KB-P04T01, KB-P04T03 and 01GR0471 to E.E.W. and U.S.), Deutsche Forschungsgemeinschaft (SFB 577 and SFB618 to E.E.W.), the University of Ghent and the “Fonds Wetenschappelijk Onderzoek–Vlaanderen” (FWO-V) G.0031.06 (GOA12051401 to J. Tavernier) and the National Cancer Institute of Canada (to C.B.). I.L. is a postdoctoral fellow with the FWO-V. M.V. is a “Chercheur Qualifié Honoraire” from the Fonds de la Recherche Scientifique (French Community of Belgium).
Author information
Authors and Affiliations
Contributions
K.V., J.-F.R., A. Vazquez, U.S., I.L., J. Tavernier, E.E.W., A.-L.B. and M.V. conceived the project. K.V., J.-F.R., A. Vazquez, U.S. and I.L. coordinated the experiments and data analyses. J.-F.R., U.S., T.H.-K., M.Z., X.X., K.H., F.G., J.M.S., P.B., H.Y., S.C., C.S., E.D., J. Timm, K.R. and C.B. conducted the Y2H experiments. J.-F.R., T.H.-K. and C.S. conducted the high-throughput ORF cloning for MAPPIT experiments. I.L. and A.-S.d.S. conducted the MAPPIT experiments. K.V., A. Vazquez, T.H., K.-I.G., M.A.Y., A. Vinayagam, N.S., N.K., C.L., M.L. and F.P.R. conducted the computational and statistical analyses. M.E.C., A.S., H.B., J.-F.R. and K.V. conducted the literature-curated interaction recuration analyses. D.S., A.D. and R.R.M. provided laboratory support. K.V., J.-F.R., A. Vazquez, U.S., I.L., M.E.C., D.E.H., J. Tavernier, E.E.W., A.-L.B. and M.V. wrote the manuscript. D.E.H, J. Tavernier, E.E.W., A.-L.B. and M.V. codirected the project.
Corresponding authors
Supplementary information
Supplementary Text and Figures
Supplementary Figure 1; Supplementary Tables 2,3,5; Supplementary Data 1–4; Supplementary Methods (PDF 2639 kb)
Supplementary Table 1
List of interactions in various datasets used in pair wise test experiments using MAPPIT and Y2H-CCSB assays (XLS 144 kb)
Supplementary Table 4
Scores for the Y2H-CCSB and MAPPIT experiments on the hsPRS-v1 and hsRRS-v1 to compute assay sensitivity and background positive rate and scores on subsets of the LC, MDC-HI1 and CCSB-HI1 interaction datasets (XLS 78 kb)
Supplementary Table 6
Identity of the ORFs making up the Y2H-CCSB repeat screens (XLS 396 kb)
Supplementary Table 7
Interactions found in the Y2H-CCSB repeat screens (XLS 69 kb)
Supplementary Table 8
Interactions found in the Y2H-CCSB repeat screens reported according to MIMIX specifications (XLS 207 kb)
Supplementary Table 9
Identity of the ORFs making up the MDC-HI1 search space (space II) (XLS 159 kb)
Rights and permissions
About this article
Cite this article
Venkatesan, K., Rual, JF., Vazquez, A. et al. An empirical framework for binary interactome mapping. Nat Methods 6, 83–90 (2009). https://doi.org/10.1038/nmeth.1280
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nmeth.1280
This article is cited by
-
A binary interaction map between turnip mosaic virus and Arabidopsis thaliana proteomes
Communications Biology (2023)
-
The protein interactome of the citrus Huanglongbing pathogen Candidatus Liberibacter asiaticus
Nature Communications (2023)
-
Speos: an ensemble graph representation learning framework to predict core gene candidates for complex diseases
Nature Communications (2023)
-
Symbiont-host interactome mapping reveals effector-targeted modulation of hormone networks and activation of growth promotion
Nature Communications (2023)
-
Next-generation large-scale binary protein interaction network for Drosophila melanogaster
Nature Communications (2023)