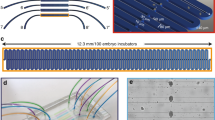
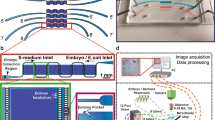
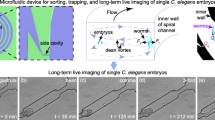
Abstract
Microscopy, phenotyping and visual screens are frequently applied to model organisms in combination with genetics. Although widely used, these techniques for multicellular organisms have mostly remained manual and low-throughput. Here we report the complete automation of sample handling, high-resolution microscopy, phenotyping and sorting of Caenorhabditis elegans. The engineered microfluidic system, coupled with customized software, has enabled high-throughput, high-resolution microscopy and sorting with no human intervention and may be combined with any microscopy setup. The microchip is capable of robust local temperature control, self-regulated sample-loading and automatic sample-positioning, while the integrated software performs imaging and classification of worms based on morphological and intensity features. We demonstrate the ability to perform sensitive and quantitative screens based on cellular and subcellular phenotypes with over 95% accuracy per round and a rate of several hundred worms per hour. Screening time can be reduced by orders of magnitude; moreover, screening is completely automated.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Fire, A. et al. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391, 806–811 (1998).
Sonnichsen, B. et al. Full-genome RNAi profiling of early embryogenesis in Caenorhabditis elegans. Nature 434, 462–469 (2005).
Kamath, R.S. & Ahringer, J. Genome-wide RNAi screening in Caenorhabditis elegans. Methods 30, 313–321 (2003).
Sieburth, D. et al. Systematic analysis of genes required for synapse structure and function. Nature 436, 510–517 (2005).
Hunt-Newbury, R. et al. High-throughput in vivo analysis of gene expression in caenorhabditis elegans. PLoS Biol. 5, e237 (2007).
Piano, F., Gunsalus, K.C., Hill, D.E. & Vidal, M. C. elegans network biology: a beginning. in WormBook (ed. The C. elegans Research Community). Published online 21 August, 2006 (doi:10.1895/wormbook.1.118.1).
Dupuy, D. et al. Genome-scale analysis of in vivo spatiotemporal promoter activity in Caenorhabditis elegans. Nat. Biotechnol. 25, 663–668 (2007).
Mango, S.E. A green light to expression in time and space. Nat. Biotechnol. 25, 645–646 (2007).
Duverger, Y. et al. A semi-automated high-throughput approach to the generation of transposon insertion mutants in the nematode Caenorhabditis elegans. Nucleic Acids Res. 35, e11 (2007).
Troemel, E.R., Sagasti, A. & Bargmann, C.I. Lateral signaling mediated by axon contact and calcium entry regulates asymmetric odorant receptor expression in C. elegans. Cell 99, 387–398 (1999).
Klassen, M.P. & Shen, K. Wnt signaling positions neuromuscular connectivity by inhibiting synapse formation in C. elegans. Cell 130, 704–716 (2007).
Zhao, H. & Nonet, M.L. A retrograde signal is involved in activity-dependent remodeling at a C. elegans neuromuscular junction. Development 127, 1253–1266 (2000).
Zhen, M. & Jin, Y. The liprin protein SYD-2 regulates the differentiation of presynaptic termini in C. elegans. Nature 401, 371–375 (1999).
Schaefer, A.M., Hadwiger, G.D. & Nonet, M.L. rpm-1, a conserved neuronal gene that regulates targeting and synaptogenesis in C. elegans. Neuron 26, 345–356 (2000).
Shen, K. & Bargmann, C.I. The immunoglobulin superfamily protein SYG-1 determines the location of specific synapses in C. elegans. Cell 112, 619–630 (2003).
Hobert, O., Johnston, R.J. & Chang, S. Left-right asymmetry in the nervous system: The Caenorhabditis elegans model. Nat. Rev. Neurosci. 3, 629–640 (2002).
Lucchetta, E.M., Lee, J.H., Fu, L.A., Patel, N.H. & Ismagilov, R.F. Dynamics of Drosophila embryonic patterning network perturbed in space and time using microfluidics. Nature 434, 1134–1138 (2005).
Funfak, A., Brösing, A., Brand, M. & Köhler, J.M. Micro fluid segment technique for screening and development studies on Danio rerio embryos. Lab Chip 7, 1132–1138 (2007).
Gray, J.M. et al. Oxygen sensation and social feeding mediated by a C-elegans guanylate cyclase homologue. Nature 430, 317–322 (2004).
Zhang, Y., Lu, H. & Bargmann, C.I. Pathogenic bacteria induce aversive olfactory learning in Caenorhabditis elegans. Nature 438, 179–184 (2005).
Chronis, N., Zimmer, M. & Bargmann, C.I. Microfluidics for in vivo imaging of neuronal and behavioral activity in Caenorhabditis elegans. Nat. Methods 4, 727–731 (2007).
Heng, X. et al. Optofluidic microscopy—a method for implementing a high resolution optical microscope on a chip. Lab Chip 6, 1274–1276 (2006).
Hulme, S.E., Shevkoplyas, S.S., Apfeld, J., Fontana, W. & Whitesides, G.M. A microfabricated array of clamps for immobilizing and imaging C. elegans. Lab Chip 7, 1515–1523 (2007).
Rohde, C.B., Zeng, F., Gonzalez-Rubio, R., Angel, M. & Yanik, M.F. Microfluidic system for on-chip high-throughput whole-worm sorting and screening at subcellular resolution. Proc. Natl. Acad. Sci. USA 104, 13891–13895 (2007).
Guo, S.X. et al. Femtosecond laser nanoaxotomy lab-on-a-chip for in vivo nerve regeneration studies. Nat. Methods 5, 531–533 (2008).
Chang, A.J., Chronis, N., Karow, D.S., Marletta, M.A. & Bargmann, C.I. A distributed chemosensory circuit for oxygen preference in C. elegans. PLoS Biol. 4, e274 (2006).
Grill, B. et al. C. elegans RPM-1 regulates axon termination and synaptogenesis through the Rab GEF GLO-4 and the Rab GTPase GLO-1. Neuron 55, 587–601 (2007).
Dittman, J.S. & Kaplan, J.M. Factors regulating the abundance and localization of synaptobrevin in the plasma membrane. Proc. Natl. Acad. Sci. USA 103, 11399–11404 (2006).
Brenner, S. The genetics of Caenorhabditis elegans. Genetics 77, 71–94 (1974).
Unger, M.A., Chou, H.P., Thorsen, T., Scherer, A. & Quake, S.R. Monolithic microfabricated valves and pumps by multilayer soft lithography. Science 288, 113–116 (2000).
Acknowledgements
We acknowledge US National Science Foundation (DBI-0649833) and National Institutes of Health (NS058465) for funding, Caenorhabditis Genetics Center, Y. Jin (University of California San Diego), and C.I. Bargmann (Rockefeller University) for providing strains, H. Brown and Y. Jin for sharing unpublished observations, J. Stirman for technical assistance, and V. Breedveld, R. Butera, T. Streelman and Y. Thio for commenting on the manuscript. M.M.C. is a National Science Foundation graduate fellow.
Author information
Authors and Affiliations
Contributions
K.C., M.M.C. and H.L. designed the experiments. K.C. fabricated the devices, M.M.C. implemented the software, and K.C. and M.M.C. conducted the experiments. K.C., M.M.C. and H.L. analyzed the data and wrote the paper.
Corresponding author
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1-7, Supplementary Methods (PDF 1184 kb)
Supplementary Video 1
No cooling. This movie shows a nematode being imaged at high magnification (100X) without cooling. A pressure gradient is applied to the worm to push it against the side channels in an attempt to keep the animal from moving. As can be easily seen, the worm is still able to move and rotate significantly. (MOV 2199 kb)
Supplementary Video 2
Cooling. This movie shows a nematode being imaged at high magnification (100X) with cooling applied. Unlike Video 1, no pressure gradient is applied to the animal to keep it in place. With cooling alone the animal remains perfectly still for the entire duration of the imaging process. (MOV 2179 kb)
Supplementary Video 3
Routing. This movie shows worms entering the observation chamber and then being routed to either the top or bottom outlet. This demonstrates the routing process of worms during sorting. (MOV 1490 kb)
Supplementary Video 4
Loading regulator. This movie shows the functioning loading regulator. When a worm is positioned inside the observation chamber, the pressure drop across the worm positioned behind the loading regulator is too small to push it into the imaging area. (MOV 1039 kb)
Supplementary Video 5
AWC 1 ON. Representative z-stack of strain CX3695 at 100x on-chip immobilized using cooling showing only one AWC cell body and dendritic/axonal processes. (MOV 498 kb)
Supplementary Video 6
AWC 2 ON. Representative z-stack of strain CX3940 at 100x on-chip immobilized using cooling showing two AWC cell bodies and dendritic/axonal processes. (MOV 736 kb)
Supplementary Video 7
CZ5261. Representative z-stack of strain CZ5261 at 100x on-chip immobilized using cooling. (MOV 648 kb)
Supplementary Video 8
CZ5264. Representative z-stack of strain CZ5264 at 100x on-chip immobilized using cooling. (MOV 1494 kb)
Rights and permissions
About this article
Cite this article
Chung, K., Crane, M. & Lu, H. Automated on-chip rapid microscopy, phenotyping and sorting of C. elegans. Nat Methods 5, 637–643 (2008). https://doi.org/10.1038/nmeth.1227
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nmeth.1227
This article is cited by
-
A robot-assisted acoustofluidic end effector
Nature Communications (2022)
-
Microfluidics for understanding model organisms
Nature Communications (2022)
-
C. elegans: A biosensor for host–microbe interactions
Lab Animal (2021)
-
Pressure measurement methods in microchannels: advances and applications
Microfluidics and Nanofluidics (2021)
-
Microfluidics for interrogating live intact tissues
Microsystems & Nanoengineering (2020)