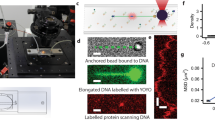
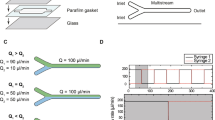
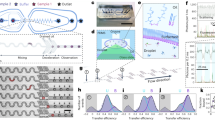
Abstract
Microfluidic flow cells are used in single-molecule experiments, enabling measurements to be made with high spatial and temporal resolution. We discuss the fundamental processes affecting flow cell operation and describe the flow cells in use at present for studying the interaction of optically trapped or mechanically isolated, single DNA molecules with proteins. To assist the experimentalist in flow cell selection, we review the construction techniques and materials used to fabricate both single- and multiple-channel flow cells and the advantages of each design for different experiments.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Bianco, P.R. et al. Processive translocation and DNA unwinding by individual RecBCD enzyme molecules. Nature 409, 374–378 (2001).
Perkins, T.T. et al. Forward and reverse motion of single RecBCD molecules on DNA. Biophys. J. 86, 1640–1648 (2004).
Handa, N., Bianco, P.R., Baskin, R.J. & Kowalczykowski, S. Direct visualization of RecBCD movement reveals cotranslocation of the RecD motor after χ recognition. Mol. Cell 17, 745–750 (2005).
Spies, M. et al. A molecular throttle: the recombination hotspot chi controls DNA translocation by the RecBCD helicase. Cell 114, 647–654 (2003).
Chemla, Y.R. et al. Mechanism of force generation of a viral DNA packaging motor. Cell 122, 683–692 (2005).
Grayson, P., Han, L., Winther, T. & Phillips, R. Real-time observations of single bacteriophage lambda DNA ejections in vitro. Proc. Natl. Acad. Sci. USA 104, 14652–14657 (2007).
Amitani, I., Baskin, R.J. & Kowalczykowski, S.C. Visualization of Rad54, a chromatin remodeling protein, translocating on single DNA molecules. Mol. Cell 23, 143–148 (2006).
Bianco, P.R., Bradfield, J.J., Castanza, L.R. & Donnelly, A.N. Rad54 oligomers translocate and cross-bridge double-stranded DNA to stimulate synapsis. J. Mol. Biol. 374, 618–640 (2007).
van Oijen, A.M. et al. Single-molecule kinetics of lambda exonuclease reveal base dependence and dynamic disorder. Science 301, 1235–1238 (2003).
Wuite, G.J.L., Smith, S.B., Young, M., Keller, D. & Bustamante, C. Single-molecule studies of the effect of template tension on T7 DNA polymerase activity. Nature 404, 103–106 (2000).
Luo, G., Wang, M., Konigsberg, W.H. & Xie, X.S. Single-molecule and ensemble fluorescence assays for a functionally important conformational change in T7 DNA polymerase. Proc. Natl. Acad. Sci. USA 104, 12610–12615 (2007).
Tanner, N.A. et al. Single-molecule studies of fork dynamics in Escherichia coli DNA replication. Nat. Struct. Mol. Biol. 15, 170–176 (2008).
Harada, Y. et al. Single-molecule imaging of RNA polymerase-DNA interactions in real time. Biophys. J. 76, 709–715 (1999).
Davenport, R.J., Wuite, G.J., Landick, R. & Bustamante, C. Single-molecule study of transcriptional pausing and arrest by E. coli RNA polymerase. Science 287, 2497–2500 (2000).
Cheng, W., Dumont, S., Tinoco, I. Jr. & Bustamante, C. NS3 helicase actively separates RNA strands and senses sequence barriers ahead of the opening fork. Proc. Natl. Acad. Sci. USA 104, 13954–13959 (2007).
Dumont, S. et al. RNA translocation and unwinding mechanism of HCV NS3 helicase and its coordination by ATP. Nature 439, 105–108 (2006).
Shaqfeh, E.S. The dynamics of single-molecule DNA in flow. J. Non-Newt. Fluid Mech. 130, 1–28 (2005).
Bustamante, C. Unfolding single RNA molecules: bridging the gap between equilibrium and non-equilibrium statistical thermodynamics. Q. Rev. Biophys. 38, 291–301 (2005).
Brewer, L.R., Corzett, M. & Balhorn, R. Protamine-induced condensation and decondensation of the same DNA molecule. Science 286, 120–123 (1999).
Bennink, M.L. et al. Single-molecule manipulation of double-stranded DNA using optical tweezers: interaction studies of DNA with RecA and YOYO-1. Cytometry 36, 200–208 (1999).
Bennink, M.L. et al. Unfolding individual nucleosomes by stretching single chromatin fibers with optical tweezers. Nat. Struct. Biol. 8, 606–610 (2001).
Bustamante, C., Bryant, Z. & Smith, S.B. Ten years of tension: single-molecule DNA mechanics. Nature 421, 423–427 (2003).
Greene, E.C. & Mizuuchi, K. Direct observation of single MuB polymers: evidence for a DNA-dependent conformational change for generating an active target complex. Mol. Cell 9, 1079–1089 (2002).
Dame, R.T., Noom, M.C. & Wuite, G.J. Bacterial chromatin organization by H-NS protein unravelled using dual DNA manipulation. Nature 444, 387–390 (2006).
Galletto, R., Amitani, I., Baskin, R.J. & Kowalczykowski, S.C. Direct observation of individual RecA filaments assembling on single DNA molecules. Nature 443, 875–878 (2006).
Tan, X., Mizuuchi, M. & Mizuuchi, K. DNA transposition target immunity and the determinants of the MuB distribution patterns on DNA. Proc. Natl. Acad. Sci. USA 104, 13925–13929 (2007).
Squires, T.M. & Quake, S.R. Microfluidics: fluid physics at the nanoliter scale. Rev. Mod. Phys. 77, 977–1026 (2005).
Weibel, D.B. & Whitesides, G.M. Applications of microfluidics in chemical biology. Curr. Opin. Chem. Biol. 10, 584–591 (2006).
Levin, S. & Tawil, G. Effect of surfactants on the diffusion coefficients of proteins, measured by analytical SPLITT fractionation (ASF) in the diffusion mode. J. Pharm. Biomed. Anal. 12, 499–507 (1994).
Fuh, C.B., Levin, S. & Giddings, J.C. Rapid diffusion coefficient measurements using analytical SPLITT fractionation: application to proteins. Anal. Biochem. 208, 80–87 (1993).
Ismagilov, R.F., Stroock, A.D., Kenis, P.J.A., Whitesides, G.M. & Stone, H.A. Experimental and theoretical scaling laws for transverse diffusive broadening in two-phase laminar flows in microchannels. Appl. Phys. Lett. 76, 2376–2378 (2000).
Kamholz, A.E. & Yager, P. Theoretical analysis of molecular diffusion in pressure-driven laminar flow in microfluidic channels. Biophys. J. 80, 155–160 (2001).
Kamholz, A.E., Schilling, E.A. & Yager, P. Optical measurement of transverse molecular diffusion in a microchannel. Biophys. J. 80, 1967–1972 (2001).
Lima, R., Wada, S., Takeda, M., Tsubota, K. & Yamaguchi, T. In vitro confocal micro-PIV measurements of blood flow in a square microchannel: the effect of the haematocrit on instantaneous velocity profiles. J. Biomech. 40, 2752–2757 (2007).
Hong, J.W. & Quake, S.R. Integrated nanoliter systems. Nat. Biotechnol. 21, 1179–1183 (2003).
Psaltis, D., Quake, S.R. & Yang, C. Developing optofluidic technology through the fusion of microfluidics and optics. Nature 442, 381–386 (2006).
Weibel, D.B., Diluzio, W.R. & Whitesides, G.M. Microfabrication meets microbiology. Nat. Rev. Microbiol. 5, 209–218 (2007).
Wabuyele, M.B., Ford, S.M., Stryjewski, W., Barrow, J. & Soper, S.A. Single molecule detection of double-stranded DNA in poly(methylmethacrylate) and polycarbonate microfluidic devices. Electrophoresis 22, 3939–3948 (2001).
McDonald, J.C. & Whitesides, G.M. Poly(dimethylsiloxane) as a material for fabricating microfluidic devices. Acc. Chem. Res. 35, 491–499 (2002).
Qi, S. et al. Microfluidic devices fabricated in poly(methyl methacrylate) using hot-embossing with integrated sampling capillary and fiber optics for fluorescence detection. Lab Chip 2, 88–95 (2002).
Ladoux, B., Quivy, J.P., Doyle, P.S., Almouzni, G. & Viovy, J.L. Direct imaging of single-molecules: from dynamics of a single DNA chain to the study of complex DNA-protein interactions. Sci. Prog. 84, 267–290 (2001).
Merenda, F. et al. Refractive multiple optical tweezers for parallel biochemical analysis in micro-fluidics. in Proc. SPIE, 6483, (eds. Andrews, D.L., Galves, E.J. & Nienhuis, G.) 64830A (SPIE, Bellingham, Washington, USA, 2007).
Quake, S.R. & Scherer, A. From micro- to nanofabrication with soft materials. Science 290, 1536–1540 (2000).
De Jong, J., Lammertink, R.G. & Wessling, M. Membranes and microfluidics: a review. Lab Chip 6, 1125–1139 (2006).
Ng, J.M., Gitlin, I., Stroock, A.D. & Whitesides, G.M. Components for integrated poly(dimethylsiloxane) microfluidic systems. Electrophoresis 23, 3461–3473 (2002).
Wuite, G.J.L., Davenport, R.J., Rappaport, A. & Bustamante, C. An integrated laser trap/flow control video microscope for the study of single biomolecules. Biophys. J. 79, 1155–1167 (2000).
Noom, M.C., van den Broek, B., van Mameren, J. & Wuite, G.J.L. Visualizing single DNA-bound proteins using DNA as a scanning probe. Nat. Methods 4, 1031–1036 (2007).
Mijatovic, D., Eijkel, J.C. & van den Berg, A. Technologies for nanofluidic systems: top-down vs. bottom-up—a review. Lab Chip 5, 492–500 (2005).
Rusu, C. et al. Direct integration of micromachined pipettes in a flow channel for single DNA molecule study by optical tweezers. J. Microelectromech. Syst. 10, 238–244 (2001).
Takayama, S. et al. Patterning cells and their environments using multiple laminar fluid flows in capillary networks. Proc. Natl. Acad. Sci. USA 96, 5545–5548 (1999).
Kenis, P.J. et al. Fabrication inside microchannels using fluid flow. Acc. Chem. Res. 33, 841–847 (2000).
Sia, S.K. & Whitesides, G.M. Microfluidic devices fabricated in poly(dimethylsiloxane) for biological studies. Electrophoresis 24, 3563–3576 (2003).
Kim, S., Blainey, P.C., Schroeder, C.M. & Xie, X.S. Multiplexed single-molecule assay for enzymatic activity on flow-stretched DNA. Nat. Methods 4, 397–399 (2007).
Tanaka, H., Ishijima, A., Honda, M., Saito, K. & Yanagida, T. Orientation dependence of displacements by a single one-headed myosin relative to the actin filament. Biophys. J. 75, 1886–1894 (1998).
Leuba, S.H., Bennink, M.L. & Zlatanova, J. Single-molecule analysis of chromatin. Methods Enzymol. 376, 73–105 (2004).
Zheng, H., Tomschik, M., Zlatanova, J. & Leuba, S.H. Evanescent field fluorescence microscopy for analysis of protein/DNA interactions at the single-molecule level. in Protein Protein Interactions, a Molecular Cloning Manual. (eds. Golemis, E. & Adams, P.) 429–444 (Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York, USA, 2005).
Lee, J.B. et al. DNA primase acts as a molecular brake in DNA replication. Nature 439, 621–624 (2006).
Brewer, L., Corzett, M. & Balhorn, R. Condensation of DNA by spermatid basic nuclear proteins. J. Biol. Chem. 277, 38895–38900 (2002).
Brewer, L., Corzett, M., Lau, E.Y. & Balhorn, R. Dynamics of protamine 1 binding to single DNA molecules. J. Biol. Chem. 278, 42403–42408 (2003).
Moffitt, J.R., Chemla, Y.R., Izhaky, D. & Bustamante, C. Differential detection of dual traps improves the spatial resolution of optical tweezers. Proc. Natl. Acad. Sci. USA 103, 9006–9011 (2006).
Acknowledgements
We would like to thank C. Bustamante, Kiyoshi Mizuuchi, G. Wuite, H. Gao and M. Gao for critical reading of the manuscript. We would also like to thank K. Mizuuchi and G. Wuite for sharing unpublished flow cell parameters and flow velocities. Funding for the experimental work in the Brewer group was provided by US National Institutes of Health grant HD01387 to L.R.B. Funding for the experimental work in the Bianco group was from US National Institutes of Health grant GM66831 and Susan G. Komen Breast Cancer Foundation grant BCTR0601350 to P.R.B.
Author information
Authors and Affiliations
Corresponding authors
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–2, Supplementary Table 1 (PDF 258 kb)
Rights and permissions
About this article
Cite this article
Brewer, L., Bianco, P. Laminar flow cells for single-molecule studies of DNA-protein interactions. Nat Methods 5, 517–525 (2008). https://doi.org/10.1038/nmeth.1217
Published:
Issue Date:
DOI: https://doi.org/10.1038/nmeth.1217
This article is cited by
-
Characterizing microfluidic approaches for a fast and efficient reagent exchange in single-molecule studies
Scientific Reports (2020)
-
The advantages of microfluidics to study actin biochemistry and biomechanics
Journal of Muscle Research and Cell Motility (2020)
-
Additive manufacturing of laminar flow cells for single-molecule experiments
Scientific Reports (2019)
-
Exploring protein-DNA interactions in 3D using in situ construction, manipulation and visualization of individual DNA dumbbells with optical traps, microfluidics and fluorescence microscopy
Nature Protocols (2013)
-
Manufacturing process and thermal characterization of a fast temperature switching microdevice for real-time biological experiments
Microsystem Technologies (2010)