Abstract
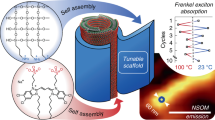
Natural light-harvesting systems spatially organize densely packed chromophore aggregates using rigid protein scaffolds to achieve highly efficient, directed energy transfer. Here, we report a synthetic strategy using rigid DNA scaffolds to similarly program the spatial organization of densely packed, discrete clusters of cyanine dye aggregates with tunable absorption spectra and strongly coupled exciton dynamics present in natural light-harvesting systems. We first characterize the range of dye-aggregate sizes that can be templated spatially by A-tracts of B-form DNA while retaining coherent energy transfer. We then use structure-based modelling and quantum dynamics to guide the rational design of higher-order synthetic circuits consisting of multiple discrete dye aggregates within a DX-tile. These programmed circuits exhibit excitonic transport properties with prominent circular dichroism, superradiance, and fast delocalized exciton transfer, consistent with our quantum dynamics predictions. This bottom-up strategy offers a versatile approach to the rational design of strongly coupled excitonic circuits using spatially organized dye aggregates for use in coherent nanoscale energy transport, artificial light-harvesting, and nanophotonics.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Scholes, G. D., Fleming, G. R., Olaya-Castro, A. & van Grondelle, R. Lessons from nature about solar light harvesting. Nat. Chem. 3, 763–774 (2011).
Aida, T., Meijer, E. W. & Stupp, S. I. Functional supramolecular polymers. Science 335, 813–817 (2012).
Stefik, M. et al. Block copolymer self-assembly for nanophotonics. Chem. Soc. Rev. 44, 5076–5091 (2015).
Son, H. J. et al. Light-harvesting and ultrafast energy migration in porphyrin-based metal-organic frameworks. J. Am. Chem. Soc. 135, 862–869 (2013).
Park, H. et al. Enhanced energy transport in genetically engineered excitonic networks. Nat. Mater. 15, 211–216 (2016).
Hannestad, J. K., Sandin, P. & Albinsson, B. Self-assembled DNA photonic wire for long-range energy transfer. J. Am. Chem. Soc. 130, 15889–15895 (2008).
Stein, I. H., Steinhauer, C. & Tinnefeld, P. Single-molecule four-color FRET visualizes energy-transfer paths on DNA origami. J. Am. Chem. Soc. 133, 4193–4195 (2011).
Dutta, P. K. et al. DNA-directed artificial light-harvesting antenna. J. Am. Chem. Soc. 133, 11985–11993 (2011).
Pan, K., Boulais, É., Yang, L. & Bathe, M. Structure-based model for light-harvesting properties of nucleic acid nanostructures. Nucleic Acids Res. 42, 2159–2170 (2014).
Buckhout-White, S. et al. Assembling programmable FRET-based photonic networks using designer DNA scaffolds. Nat. Commun. 5, 5615 (2014).
Hemmig, E. A. et al. Programming light-harvesting efficiency using DNA origami. Nano Lett. 16, 2369–2374 (2016).
Melinger, J. S. et al. FRET from multiple pathways in fluorophore-labeled DNA. ACS Photon. 3, 659–669 (2016).
Haedler, A. T. et al. Long-range energy transport in single supramolecular nanofibres at room temperature. Nature 523, 196–199 (2015).
Hannah, K. C. & Armitage, B. A. DNA-templated assembly of helical cyanine dye aggregates: a supramolecular chain polymerization. Acc. Chem. Res. 37, 845–853 (2004).
Asanuma, H., Fujii, T., Kato, T. & Kashida, H. Coherent interactions of dyes assembled on DNA. J. Photochem. Photobiol. C 13, 124–135 (2012).
Probst, M., Langenegger, S. M. & Häner, R. A modular LHC built on the DNA three-way junction. Chem. Commun. 50, 159–161 (2014).
Vybornyi, M., Nussbaumer, A. L., Langenegger, S. M. & Häner, R. Assembling multiporphyrin stacks inside the DNA double helix. Bioconjug. Chem. 25, 1785–1793 (2014).
Seeman, N. C. Nucleic acid junctions and lattices. J. Theor. Biol. 99, 237–247 (1982).
Fassioli, F., Dinshaw, R., Arpin, P. C. & Scholes, G. D. Photosynthetic light harvesting: excitons and coherence. J. R. Soc. Interface 11, 20130901 (2014).
Cunningham, P. D. et al. Optical determination of the electronic coupling and intercalation geometry of thiazole orange homodimer in DNA. J. Chem. Phys. 147, 055101 (2017).
Nicoli, F. et al. Proximity-induced H-aggregation of cyanine dyes on DNA-duplexes. J. Phys. Chem. A 120, 9941–9947 (2016).
Rothemund, P. W. K. Folding DNA to create nanoscale shapes and patterns. Nature 440, 297–302 (2006).
Han, D. et al. DNA origami with complex curvatures in three-dimensional space. Science 332, 342–346 (2011).
Ke, Y., Ong, L. L., Shih, W. M. & Yin, P. Three-dimensional structures self-assembled from DNA bricks. Science 338, 1177–1183 (2012).
Han, D. et al. DNA gridiron nanostructures based on four-arm junctions. Science 339, 1412–1415 (2013).
Zhang, F. et al. Complex wireframe DNA origami nanostructures with multi-arm junction vertices. Nat. Nanotech. 10, 779–784 (2015).
Benson, E. et al. DNA rendering of polyhedral meshes at the nanoscale. Nature 523, 441–444 (2015).
Veneziano, R. et al. Designer nanoscale DNA assemblies programmed from the top down. Science 352, 1534 (2016).
Norden, B. & Tjerneld, F. Optical studies on complexes between DNA and pseudoisocyanine. Biophys. Chem. 6, 31–45 (1976).
Meier, T., Chernyak, V. & Mukamel, S. Multiple exciton coherence sizes in photosynthetic antenna complexes viewed by pump-probe spectroscopy. J. Phys. Chem. B 101, 7332–7342 (1997).
Jones, M. R., Seeman, N. C. & Mirkin, C. A. Programmable materials and the nature of the DNA bond. Science 347, 1260901 (2015).
Wang, T. et al. Design and characterization of 1D nanotubes and 2D periodic arrays self-assembled from DNA multi-helix bundles. J. Am. Chem. Soc. 134, 1606–1616 (2012).
Kuzyk, A. et al. DNA-based self-assembly of chiral plasmonic nanostructures with tailored optical response. Nature 483, 311–314 (2012).
Sun, W. et al. Casting inorganic structures with DNA molds. Science 346, 1258361 (2014).
Helmi, S., Ziegler, C., Kauert, D. J. & Seidel, R. Shape-controlled synthesis of gold nanostructures using DNA origami molds. Nano Lett. 14, 6693–6698 (2014).
Roller, E.-M. et al. Plasmon–exciton coupling using DNA templates. Nano Lett. 16, 5962–5966 (2016).
Tian, Y. et al. Lattice engineering through nanoparticle-DNA frameworks. Nat. Mater. 15, 654–661 (2016).
Olivares-Amaya, R. et al. Accelerated computational discovery of high-performance materials for organic photovoltaics by means of cheminformatics. Energy Environ. Sci. 4, 4849–4861 (2011).
Kim, D.-N., Kilchherr, F., Dietz, H. & Bathe, M. Quantitative prediction of 3D solution shape and flexibility of nucleic acid nanostructures. Nucleic Acids Res. 40, 2862–2868 (2011).
Castro, C. E. et al. A primer to scaffolded DNA origami. Nat. Methods 8, 221–229 (2011).
Pan, K. et al. Lattice-free prediction of three-dimensional structure of programmed DNA assemblies. Nat. Commun. 5, 5578 (2014).
Valiev, M. et al. NWChem: a comprehensive and scalable open-source solution for large scale molecular simulations. Comput. Phys. Commun. 181, 1477–1489 (2010).
Combs, S. A. et al. Small-molecule ligand docking into comparative models with Rosetta. Nat. Protoc. 8, 1277–1298 (2013).
Foloppe, N. & MacKerell, A. D. All-atom empirical force field for nucleic acids: I. Parameter optimization based on small molecule and condensed phase macromolecular target data. J. Comput. Chem. 21, 86–104 (2000).
Vanommeslaeghe, K. et al. CHARMM general force field: a force field for drug-like molecules compatible with the CHARMM all-atom additive biological force fields. J. Comput. Chem. 31, 671–690 (2010).
Phillips, J. C. et al. Scalable molecular dynamics with NAMD. J. Comput. Chem. 26, 1781–1802 (2005).
Stone, J. E. et al. Accelerating molecular modeling applications with graphics processors. J. Comput. Chem. 28, 2618–2640 (2007).
Ishizaki, A. & Fleming, G. R. On the adequacy of the Redfield equation and related approaches to the study of quantum dynamics in electronic energy transfer. J. Chem. Phys. 130, 234110 (2009).
Zhong, X. & Zhao, Y. Charge carrier dynamics in phonon-induced fluctuation systems from time-dependent wavepacket diffusion approach. J. Chem. Phys. 135, 134110 (2011).
Sawaya, N. P. D. et al. Fast delocalization leads to robust long-range excitonic transfer in a large quantum chlorosome model. Nano Lett. 15, 1722–1729 (2015).
Acknowledgements
We are grateful for fruitful conversations with M. Adendorff, S. Ratanalert, T. Fujita, S. Valleau and J. Suh. Funding from the ARO MURI W911NF1210420 to M.B., N.W.W. and H.Y. is gratefully acknowledged. This work received support to M.B., G.S.S.C. and A.A.-G. from the MIT Center for Excitonics, an Energy Frontier Research Center funded by the US Department of Energy under award DE-SC0001088. M.B. additionally acknowledges funding from ONR DURIP N00014-16-1-2506 and ONR N00014-16-1-2181. A portion of the simulations was performed on Harvard University’s Odyssey cluster, supported by the Research Computing Group of the FAS Division of Science. Some of the hardware used was provided by Harvard University’s CUDA Center of Excellence (CCOE) Program, sponsored by NVIDIA. N.S. acknowledges funding from the Smith Family Graduate Science and Engineering Fellowship. The Biophysical Instrumentation Facility for the Study of Complex Macromolecular Systems (NSF-0070319) is gratefully acknowledged. E.B. acknowledges funding from the Natural Sciences and Engineering Research Council of Canada (NSERC).
Author information
Authors and Affiliations
Contributions
E.B. designed and performed molecular modelling and exciton transport simulations, designed and performed CD experiments, designed UV–vis and fluorescence spectroscopy experiments, and analysed the data. N.S. designed and performed the simulations for excitonic properties and exciton transport, and analysed the data. R.V. designed and performed UV–vis and CD experiments, ITC experiments, designed the fluorescence spectroscopy experiments, prepared the samples, and analysed the data. J.L.B. designed and performed quantum yield measurements to measure the coherence length and steady-state fluorescence spectroscopy to measure energy transfer efficiency. T.K. designed and performed experiments to measure energy transfer efficiencies using time-resolved fluorescence spectroscopy. G.S.S.C. designed and supervised the experiments to measure the coherence length and energy transfer efficiencies using steady-state and time-resolved spectroscopy. A.A., S.M. and S.L. designed and performed the fluorescence and pump–probe spectroscopy experiments, and analysed the data. N.W. designed and supervised the fluorescence and pump–probe spectroscopy study, and analysed the data. H.Y. supervised the fluorescence and pump–probe spectroscopy study, and analysed the data. A.A.-G. supervised the simulation study, and analysed the data. M.B. designed and supervised the overall study, and analysed the data. E.B., N.S. and M.B. wrote the manuscript. J.L.B., R.V. and S.L. edited the manuscript and all authors commented on the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
Supplementary Information (PDF 2922 kb)
Rights and permissions
About this article
Cite this article
Boulais, É., Sawaya, N., Veneziano, R. et al. Programmed coherent coupling in a synthetic DNA-based excitonic circuit. Nat. Mater. 17, 159–166 (2018). https://doi.org/10.1038/nmat5033
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nmat5033
This article is cited by
-
A framework for multiexcitonic logic
Nature Reviews Chemistry (2024)
-
Enabling programmable dynamic DNA chemistry using small-molecule DNA binders
Nature Communications (2023)
-
Interfacing DNA nanotechnology and biomimetic photonic complexes: advances and prospects in energy and biomedicine
Journal of Nanobiotechnology (2022)
-
Nanoscale 3D spatial addressing and valence control of quantum dots using wireframe DNA origami
Nature Communications (2022)
-
Rotaxane rings promote oblique packing and extended lifetimes in DNA-templated molecular dye aggregates
Communications Chemistry (2021)