Abstract
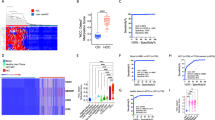
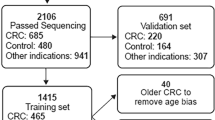
An effective blood-based method for the diagnosis and prognosis of hepatocellular carcinoma (HCC) has not yet been developed. Circulating tumour DNA (ctDNA) carrying cancer-specific genetic and epigenetic aberrations may enable a noninvasive ‘liquid biopsy’ for diagnosis and monitoring of cancer. Here, we identified an HCC-specific methylation marker panel by comparing HCC tissue and normal blood leukocytes and showed that methylation profiles of HCC tumour DNA and matched plasma ctDNA are highly correlated. Using cfDNA samples from a large cohort of 1,098 HCC patients and 835 normal controls, we constructed a diagnostic prediction model that showed high diagnostic specificity and sensitivity (P < 0.001) and was highly correlated with tumour burden, treatment response, and stage. Additionally, we constructed a prognostic prediction model that effectively predicted prognosis and survival (P < 0.001). Together, these findings demonstrate in a large clinical cohort the utility of ctDNA methylation markers in the diagnosis, surveillance, and prognosis of HCC.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Accession codes
References
Torre, L. A. et al. Global cancer statistics, 2012. CA Cancer J. Clin. 65, 87–108 (2015).
Bruix, J. & Sherman, M. AASLD Practice Guideline: Management of hepatocellular carcinoma. Hepatology 42, 1208–1236 (2005).
Johnson, P. Role of alpha - fetoprotein in the diagnosis and management of hepatocellular carcinoma. J. Gastroenterol. Hepatol. 14, S32–S36 (1999).
Stroun, M. et al. The origin and mechanism of circulating DNA. Ann. NY Acad. Sci. 906, 161–168 (2000).
Bettegowda, C. et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci. Trans. Med. 6, 224ra224 (2014).
Newman, A. M. et al. An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage. Nat. Med. 20, 548–554 (2014).
Esteller, M. Epigenetics in cancer. N. Engl. J. Med. 358, 1148–1159 (2008).
Baylin, S. B. & Jones, P. A. Epigenetic determinants of cancer. Cold Spring Harb. Perspect. Biol. 8, a019505 (2016).
Irizarry, R. A. et al. The human colon cancer methylome shows similar hypo- and hypermethylation at conserved tissue-specific CpG island shores. Nat. Genet. 41, 178–186 (2009).
Baylin, S. B. & Jones, P. A. A decade of exploring the cancer epigenome—biological and translational implications. Nat. Rev. Cancer 11, 726–734 (2011).
Board, R. E. et al. DNA methylation in circulating tumour DNA as a biomarker for cancer. Biomarker Insights 2, 307–319 (2008).
Warren, J. D. et al. Septin 9 methylated DNA is a sensitive and specific blood test for colorectal cancer. BMC Med. 9, 133 (2011).
Pishvaian, M. J. et al. A pilot study evaluating concordance between blood-based and patient-matched tumor molecular testing within pancreatic cancer patients participating in the Know Your Tumor (KYT) initiative. Oncotarget 7, 13225 (2016).
Lehmann-Werman, R. et al. Identification of tissue-specific cell death using methylation patterns of circulating DNA. Proc. Natl Acad. Sci. USA 113, 201519286 (2016).
Hannum, G. et al. Genome-wide methylation profiles reveal quantitative views of human aging rates. Mol. Cell 49, 359–367 (2013).
Smyth, G. K. Bioinformatics and Computational Biology Solutions Using R and Bioconductor 397–420 (Springer, 2005).
Benjamini, Y. & Hochberg, Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Statist. Soc. B 57, 289–300 (1995).
Reich, D. E. et al. Linkage disequilibrium in the human genome. Nature 411, 199–204 (2001).
Ardlie, K. G., Kruglyak, L. & Seielstad, M. Patterns of linkage disequilibrium in the human genome. Nat. Rev. Genet. 3, 299–309 (2002).
Hao, X. et al. DNA methylation markers for diagnosis and prognosis of common cancers. Proc. Natl Acad. Sci. USA 114, 7414–7419 (2017).
Aravanis, A. M., Lee, M. & Klausner, R. D. Next-generation sequencing of circulating tumor DNA for early cancer detection. Cell 168, 571–574 (2017).
Snyder, M. W., Kircher, M., Hill, A. J., Daza, R. M. & Shendure, J. Cell-free DNA comprises an in vivo nucleosome footprint that informs its tissues-of-origin. Cell 164, 57–68 (2016).
Singal, A. et al. Meta-analysis: surveillance with ultrasound for early-stage hepatocellular carcinoma in patients with cirrhosis. Aliment Pharmacol. Ther. 30, 37–47 (2009).
Butcher, L. M. et al. Non-CG DNA methylation is a biomarker for assessing endodermal differentiation capacity in pluripotent stem cells. Nat. Commun. 7, 10458 (2016).
Libertini, E. et al. Information recovery from low coverage whole-genome bisulfite sequencing. Nat. Commun. 7, 11306 (2016).
Burger, L., Gaidatzis, D., Schubeler, D. & Stadler, M. B. Identification of active regulatory regions from DNA methylation data. Nucleic Acids Res. 41, e155 (2013).
Guo, S. et al. Identification of methylation haplotype blocks aids in deconvolution of heterogeneous tissue samples and tumor tissue-of-origin mapping from plasma DNA. Nat. Genet. 49, 635–642 (2017).
Lencioni, R. & Llovet, J. M. Modified RECIST (mRECIST) assessment for hepatocellular carcinoma. Semin. Liver Dis. 30, 52–60 (2010).
Raoul, J. L. et al. Using modified RECIST and alpha-fetoprotein levels to assess treatment benefit in hepatocellular carcinoma. Liver Cancer 3, 439–450 (2014).
Diehl, F. et al. Circulating mutant DNA to assess tumor dynamics. Nat. Med. 14, 985–990 (2008).
Mok, T. et al. Detection and dynamic changes of EGFR mutations from circulating tumor DNA as a predictor of survival outcomes in NSCLC patients treated with first-line intercalated erlotinib and chemotherapy. Clin. Cancer Res. 21, 3196–3203 (2015).
Diaz, L. A. Jr. et al. The molecular evolution of acquired resistance to targeted EGFR blockade in colorectal cancers. Nature 486, 537–540 (2012).
Misale, S. et al. Emergence of KRAS mutations and acquired resistance to anti-EGFR therapy in colorectal cancer. Nature 486, 532–536 (2012).
Dawson, S. J. et al. Analysis of circulating tumor DNA to monitor metastatic breast cancer. N. Engl. J. Med. 368, 1199–1209 (2013).
Edge, S. B. & Compton, C. C. The American Joint Committee on Cancer: the 7th edition of the AJCC cancer staging manual and the future of TNM. Ann. Surg. Oncol. 17, 1471–1474 (2010).
Porreca, G. J. et al. Multiplex amplification of large sets of human exons. Nat. Methods 4, 931–936 (2007).
Diep, D. et al. Library-free methylation sequencing with bisulfite padlock probes. Nat. Methods 9, 270–272 (2012).
Deng, J. et al. Targeted bisulfite sequencing reveals changes in DNA methylation associated with nuclear reprogramming. Nat. Biotechnol. 27, 353–360 (2009).
Langmead, B. & Salzberg, S. L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012).
Acknowledgements
The results published here are in part based upon data generated by the TCGA Research Network: http://cancergenome.nih.gov. We thank staff at Kang Zhang and Ruihua Xu laboratories for technical assistance. This study was funded by Richard Annesser Fund, Michael Martin Fund, Dick and Carol Hertzberg Fund, SYSUCC, Xijing Hospital, and West China Hospital.
Author information
Authors and Affiliations
Contributions
W.Wei, M.K., W.Wang, H.L., K.F., W.S., S.Y., L.Z., H.Z., R.Z., Y.X., K.L., H.Cai, G.L., L.Z., R.-h.X., Z.Z., D.L., E.Z. and C.Z. performed the experiments; M.K. W.Wang, H.L., K.F., B.A.C., Q.Q., Q.Z., L.Z., R.-h.X., J.Z., X.F., J.-k.Z., Y.D., H.Carter, M.Y., W.Z., R.G. and X.H. collected and analysed the data. K.Z. and R.-h.X. conceived the project, designed the experiments, and wrote the manuscript; All authors discussed the results and reviewed the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
Supplementary Information (PDF 7011 kb)
Rights and permissions
About this article
Cite this article
Xu, Rh., Wei, W., Krawczyk, M. et al. Circulating tumour DNA methylation markers for diagnosis and prognosis of hepatocellular carcinoma. Nature Mater 16, 1155–1161 (2017). https://doi.org/10.1038/nmat4997
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nmat4997
This article is cited by
-
Novel Blood-Based Biomarkers for HCC
Current Hepatology Reports (2024)
-
Genome-wide 5-hydroxymethylcytosines in circulating cell-free DNA as noninvasive diagnostic markers for gastric cancer
Gastric Cancer (2024)
-
Update on HCC Surveillance in Patient With Hepatitis B Virus Infection With Focus on Biomarkers
Current Hepatology Reports (2024)
-
Association of Placental Tissue Metabolite Levels with Gestational Diabetes Mellitus: a Metabolomics Study
Reproductive Sciences (2024)
-
Hepatocellular carcinoma detection via targeted enzymatic methyl sequencing of plasma cell-free DNA
Clinical Epigenetics (2023)