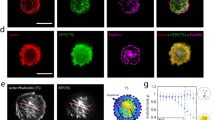
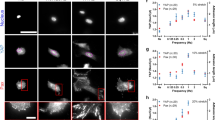
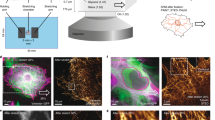
Abstract
Mechanical forces direct a host of cellular and tissue processes. Although much emphasis has been placed on cell-adhesion complexes as force sensors, the forces must nevertheless be transmitted through the cortical cytoskeleton. Yet how the actin cortex senses and transmits forces and how cytoskeletal proteins interact in response to the forces is poorly understood. Here, by combining molecular and mechanical experimental perturbations with theoretical multiscale modelling, we decipher cortical mechanosensing from molecular to cellular scales. We show that forces are shared between myosin II and different actin crosslinkers, with myosin having potentiating or inhibitory effects on certain crosslinkers. Different types of cell deformation elicit distinct responses, with myosin and α-actinin responding to dilation, and filamin mainly reacting to shear. Our observations show that the accumulation kinetics of each protein may be explained by its molecular mechanisms, and that protein accumulation and the cell’s viscoelastic state can explain cell contraction against mechanical load.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
LeDuc, P. R. & Robinson, D. N. Using lessons from cellular and molecular structures for future materials. Adv. Mater. 19, 3761–3770 (2007).
Grashoff, C. et al. Measuring mechanical tension across vinculin reveals regulation of focal adhesion dynamics. Nature 466, 263–266 (2010).
Geiger, B., Spatz, J. P. & Bershadsky, A. D. Environmental sensing through focal adhesions. Nature Rev. Mol. Cell Biol. 10, 21–33 (2009).
Tamada, M., Sheetz, M. P. & Sawada, Y. Activation of signaling cascade by cytoskeleton stretch. Dev. Cell 7, 709–718 (2004).
Chowdhury, F. et al. Material properties of the cell dictate stress-induced spreading and differentiation in embryonic stem cells. Nature Mater. 9, 82–88 (2010).
Ehrlicher, A. J., Nakamura, F., Hartwig, J. H., Weitz, D. A. & Stossel, T. P. Mechanical strain in actin networks regulates FilGAP and integrin binding to filamin A. Nature 478, 260–263 (2011).
Effler, J. C. et al. Mitosis-specific mechanosensing and contractile protein redistribution control cell shape. Curr. Biol. 16, 1962–1967 (2006).
Kee, Y-S. et al. A mechanosensory system governs myosin II accumulation in dividing cells. Mol. Biol. Cell 23, 1510–1523 (2012).
Johnson, C. P., Tang, H-Y., Carag, C., Speicher, D. W. & Discher, D. E. Forced unfolding of proteins within cells. Science 317, 663–666 (2007).
DuFort, C. C., Paszek, M. J. & Weaver, V. M. Balancing forces: Architectural control of mechanotransduction. Nature Rev. Mol. Cell Biol. 12, 308–319 (2011).
Engler, A. J., Sen, S., Sweeney, H. L. & Discher, D. E. Matrix elasticity directs stem cell lineage specification. Cell 126, 677–689 (2006).
Raab, M. et al. Crawling from soft to stiff matrix polarizes the cytoskeleton and phosphoregulates myosin-II heavy chain. J. Cell Biol. 199, 669–683 (2012).
Köhler, S., Schaller, V. & Bausch, A. R. Structure formation in active networks. Nature Mater. 10, 462–468 (2011).
Del Rio, A. et al. Stretching single talin rod molecules activates vinculin binding. Science 323, 638–641 (2009).
Gardel, M. L. et al. Elastic behavior of cross-linked and bundled actin networks. Science 304, 1301–1305 (2004).
Shin, J. H., Gardel, M. L., Mahadevan, L., Matsudaira, P. & Weitz, D. A. Relating microstructure to rheology of a bundled and cross-linked F-actin network in vitro. Proc. Natl Acad. Sci. USA 101, 9636–9641 (2004).
Chaudhuri, O., Parekh, S. H. & Fletcher, D. A. Reversible stress softening of actin networks. Nature 445, 295–298 (2007).
Koenderink, G. H. et al. An active biopolymer network controlled by molecular motors. Proc. Natl Acad. Sci. USA 106, 15192–15197 (2009).
Wagner, B., Tharmann, R., Haase, I., Fischer, M. & Bausch, A. R. Cytoskeletal polymer networks: The molecular structure of cross-linkers determines macroscopic properties. Proc. Natl Acad. Sci. USA 103, 13974–13978 (2006).
Schmoller, K. M., Lieleg, O. & Bausch, A. R. Cross-linking molecules modify composite actin networks independently. Phys. Rev. Lett. 101, 118102 (2008).
Murrell, M. et al. Spreading dynamics of biomimetic actin cortices. Biophys. J. 100, 1400–1409 (2011).
He, L., Wang, X., Tang, H. L. & Montell, D. J. Tissue elongation requires oscillating contractions of a basal actomyosin network. Nature Cell. Biol. 12, 1133–1142 (2010).
Solon, J., Kaya-Çopur, A., Colombelli, J. & Brunner, D. Pulsed forces timed by a ratchet-like mechanism drive directed tissue movement during dorsal closure. Cell 137, 1331–1342 (2009).
Bao, G. & Suresh, S. Cell and molecular mechanics of biological materials. Nature Mater. 2, 715–725 (2003).
Luo, T. et al. Understanding the cooperative interaction between myosin II and actin cross-linkers mediated by actin filaments during mechanosensation. Biophys. J. 102, 238–247 (2012).
Ren, Y. et al. Mechanosensing through cooperative interactions between myosin II and the actin crosslinker cortexillin I. Curr. Biol. 19, 1421–1428 (2009).
Uyeda, T. Q. P., Iwadate, Y., Umeki, N., Nagasaki, A. & Yumura, S. Stretching actin filaments within cells enhances their affinity for the myosin II motor domain. PLoS ONE 6, e26200 (2011).
Fernandez-Gonzalez, R., Simoes Sde, M., Roper, J. C., Eaton, S. & Zallen, J. A. Myosin II dynamics are regulated by tension in intercalating cells. Dev. Cell 17, 736–743 (2009).
Discher, D. E. & Mohandas, N. Kinematics of red cell aspiration by fluorescence-imaged microdeformation. Biophys. J. 71, 1680–1694 (1996).
Evans, E. Probing the relation between force-life-time-and chemistry in single molecular bonds. Annu. Rev. Biophys. Biomol. Struct. 30, 105–128 (2001).
Bell, G. I. Models for the specific adhesion of cells to cells. Science 200, 618–627 (1978).
Veigel, C., Molloy, J. E., Schmitz, S. & Kendrick-Jones, J. Load-dependent kinetics of force production by smooth muscle myosin measured with optical tweezers. Nature Cell Biol. 5, 980–986 (2003).
Thomas, W. E., Vogel, V. & Sokurenko, E. Biophysics of catch bonds. Annu. Rev. Biophys. 37, 399–416 (2008).
Uyeda, T. Q., Abramson, P. D. & Spudich, J. A. The neck region of the myosin motor domain acts as a lever arm to generate movement. Proc. Natl Acad. Sci. USA 93, 4459–4464 (1996).
Robinson, D. N., Kee, Y. S., Luo, T. & Surcel, A. in Comprehensive Biophysics (ed. Edward, H. E.) 48–72 (Elsevier, 2012).
Seifert, U. Rupture of multiple parallel molecular bonds under dynamic loading. Phys. Rev. Lett. 84, 2750–2753 (2000).
Erdmann, T. & Schwarz, U. S. Stability of adhesion clusters under constant force. Phys. Rev. Lett. 92, 108102 (2004).
Girard, K. D., Chaney, C., Delannoy, M., Kuo, S. C. & Robinson, D. N. Dynacortin contributes to cortical viscoelasticity and helps define the shape changes of cytokinesis. EMBO J. 23, 1536–1546 (2004).
Gerisch, G. et al. Mobile actin clusters and traveling waves in cells recovering from actin depolymerization. Biophys. J. 87, 3493–3503 (2004).
Robinson, D. N. & Spudich, J. A. Dynacortin, a genetic link between equatorial contractility and global shape control discovered by library complementation of a Dictyostelium discoideum cytokinesis mutant. J. Cell Biol. 150, 823–838 (2000).
Nakamura, F., Osborn, T. M., Hartemink, C. A., Hartwig, J. H. & Stossel, T. P. Structural basis of filamin A functions. J. Cell. Biol. 179, 1011–1025 (2007).
Meyer, R. K. & Aebi, U. Bundling of actin filaments by alpha-actinin depends on its molecular length. J. Cell Biol. 110, 2013–2024 (1990).
Hock, R. S. & Condeelis, J. S. Isolation of a 240-kilodalton actin-binding protein from Dictyostelium discoideum. J. Biol. Chem. 262, 394–400 (1987).
Ferrer, J. M. et al. Measuring molecular rupture forces between single actin filaments and actin-binding proteins. Proc. Natl Acad. Sci. USA 105, 9221–9226 (2008).
Guo, B. & Guilford, W. H. Mechanics of actomyosin bonds in different nucleotide states are tuned to muscle contraction. Proc. Natl Acad. Sci. USA. 103, 9844–9849 (2006).
Pereverzev, Y. V., Prezhdo, O. V., Forero, M., Sokurenko, E. V. & Thomas, W. E. The two-pathway model for the catch-slip transition in biological adhesion. Biophys. J. 89, 1446–1454 (2005).
Evans, E., Leung, A., Heinrich, V. & Zhu, C. Mechanical switching and coupling between two dissociation pathways in a P-selectin adhesion bond. Proc. Natl Acad. Sci. USA 101, 11282–11286 (2004).
Gimona, M., Djinovic-Carugo, K., Kranewitter, W. J. & Winder, S. J. Functional plasticity of CH domains. FEBS Lett. 513, 98–106 (2002).
Yao, N. Y. et al. Stress-enhanced gelation: A dynamic nonlinearity of elasticity. Phys. Rev. Lett. 110, 018103 (2013).
Maugis, B. et al. Dynamic instability of the intracellular pressure drives bleb-based motility. J. Cell Sci. 123, 3884–3892 (2010).
Hotulainen, P. & Lappalainen, P. Stress fibers are generated by two distinct actin assembly mechanisms in motile cells. J. Cell Biol. 173, 383–394 (2006).
Friedrich, B. M., Friedrich, E. F., Gov, N. S. & Safran, S. A. Sacromeric pattern formation by actin cluster coalescence. PLoS Comput. Biol. 8, e1002544 (2012).
Dickinson, D. J., Robinson, D. N., Nelson, W. J. & Weis, W. I. α-Catenin and IQGAP regulate myosin localization to control epithelial tube morphology in Dictyostelium morphogenesis. Dev. Cell 23, 533–546 (2012).
Chen, C-L., Wang, Y., Sesaki, H. & Iijima, M. Myosin I Links PIP3 signaling to remodeling of the actin cytoskeleton in chemotaxis. Sci. Signal. 5, ra10 (2012).
Acknowledgements
We thank P. Devreotes, M. Iijima and their laboratory members, and members of the Robinson laboratory for reagents and discussions. We thank T. Inoue, R. Rock, R. Jensen and Robinson laboratory members for comments on the manuscript. We thank dictyBase (www.dictybase.org), D. Knecht, M. Titus, G. Gerisch, T. Egelhoff and P. Steimle for reagents. We thank V. Srivastava for help with confocal imaging. This work is supported by the National Institutes of Health grants GM066817 (to D.N.R.) and GM086704 (to D.N.R. and P.A.I.).
Author information
Authors and Affiliations
Contributions
T.L. and D.N.R. conceived, designed and wrote the paper. T.L. performed the experiments and analysed the data. T.L. conducted the coarse-grained molecular simulations. K.M. and P.A.I. carried out the continuum simulations. T.L., K.M., P.A.I. and D.N.R. proposed the strain-specific molecular mechanisms for different cytoskeletal proteins and the one-dimensional retraction model. All authors discussed the results and commented on the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
Supplementary Information (PDF 5044 kb)
Supplementary Information
Supplementary Movie S1 (MPG 206 kb)
Supplementary Information
Supplementary Movie S2 (MPG 206 kb)
Supplementary Information
Supplementary Movie S3 (MPG 182 kb)
Supplementary Information
Supplementary Movie S4 (AVI 171 kb)
Supplementary Information
Supplementary Movie S5 (AVI 763 kb)
Supplementary Information
Supplementary Movie S6 (AVI 966 kb)
Supplementary Information
Supplementary Movie S7 (AVI 768 kb)
Supplementary Information
Supplementary Movie S8 (AVI 1012 kb)
Supplementary Information
Supplementary Movie S9 (AVI 990 kb)
Rights and permissions
About this article
Cite this article
Luo, T., Mohan, K., Iglesias, P. et al. Molecular mechanisms of cellular mechanosensing. Nature Mater 12, 1064–1071 (2013). https://doi.org/10.1038/nmat3772
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nmat3772
This article is cited by
-
Mechanics of the cellular microenvironment as probed by cells in vivo during zebrafish presomitic mesoderm differentiation
Nature Materials (2023)
-
Myosin and \(\upalpha\)-actinin regulation of stress fiber contractility under tensile stress
Scientific Reports (2023)
-
Interplay between mechanics and signalling in regulating cell fate
Nature Reviews Molecular Cell Biology (2022)
-
Mechanical regulation of early vertebrate embryogenesis
Nature Reviews Molecular Cell Biology (2022)
-
Weak catch bonds make strong networks
Nature Materials (2022)