Abstract
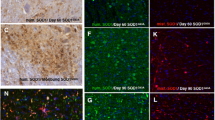
Misfolding of Cu/Zn-superoxide dismutase (SOD1) is emerging as a mechanism underlying motor neuron degeneration in individuals with amyotrophic lateral sclerosis (ALS) who carry a mutant SOD1 gene (SOD1 ALS). Here we describe a structure-guided approach to developing an antibody that specifically recognizes monomer-misfolded forms of SOD1. We raised this antibody to an epitope that is normally buried in the SOD1 native homodimer interface. The SOD1 exposed dimer interface (SEDI) antibody recognizes only those SOD1 conformations in which the native dimer is disrupted or misfolded and thereby exposes the hydrophobic dimer interface. Using the SEDI antibody, we established the presence of monomer-misfolded SOD1 in three ALS mouse models, with G37R, G85R and G93A SOD1 mutations, and in a human individual with an A4V SOD1 mutation. Despite ubiquitous expression, misfolded SOD1 was found primarily within degenerating motor neurons. Misfolded SOD1 appeared before the onset of symptoms and decreased at the end stage of the disease, concomitant with motor neuron loss.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Bruijn, L.I., Miller, T.M. & Cleveland, D.W. Unraveling the mechanisms involved in motor neuron degeneration in ALS. Annu. Rev. Neurosci. 27, 723–749 (2004).
Majoor-Krakauer, D., Willems, P.J. & Hofman, A. Genetic epidemiology of amyotrophic lateral sclerosis. Clin. Genet. 63, 83–101 (2003).
Batulan, Z. et al. High threshold for induction of the stress response in motor neurons is associated with failure to activate HSF1. J. Neurosci. 23, 5789–5798 (2003).
Urushitani, M., Kurisu, J., Tsukita, K. & Takahashi, R. Proteasomal inhibition by misfolded mutant superoxide dismutase 1 induces selective motor neuron death in familial amyotrophic lateral sclerosis. J. Neurochem. 83, 1030–1042 (2002).
Liu, J. et al. Toxicity of familial ALS-linked SOD1 mutants from selective recruitment to spinal mitochondria. Neuron 43, 5–17 (2004).
Pasinelli, P. et al. Amyotrophic lateral sclerosis-associated SOD1 mutant proteins bind and aggregate with Bcl-2 in spinal cord mitochondria. Neuron 43, 19–30 (2004).
Estevez, A.G. et al. Induction of nitric oxide-dependent apoptosis in motor neurons by zinc-deficient superoxide dismutase. Science 286, 2498–2500 (1999).
Forman, H.J. & Fridovich, I. On the stability of bovine superoxide dismutase. The effects of metals. J. Biol. Chem. 248, 2645–2649 (1973).
Borchelt, D.R. et al. Superoxide dismutase 1 with mutations linked to familial amyotrophic lateral sclerosis possesses significant activity. Proc. Natl. Acad. Sci. USA 91, 8292–8296 (1994).
Wood, J.D., Beaujeux, T.P. & Shaw, P.J. Protein aggregation in motor neurone disorders. Neuropathol. Appl. Neurobiol. 29, 529–545 (2003).
Wang, J., Xu, G. & Borchelt, D.R. High molecular weight complexes of mutant superoxide dismutase 1: age-dependent and tissue-specific accumulation. Neurobiol. Dis. 9, 139–148 (2002).
Rakhit, R. et al. Oxidation-induced misfolding and aggregation of superoxide dismutase and its implications for amyotrophic lateral sclerosis. J. Biol. Chem. 277, 47551–47556 (2002).
Rakhit, R. et al. Monomeric Cu,Zn-superoxide dismutase is a common misfolding intermediate in the oxidation models of sporadic and familial amyotrophic lateral sclerosis. J. Biol. Chem. 279, 15499–15504 (2004).
Kayed, R. et al. Common structure of soluble amyloid oligomers implies common mechanism of pathogenesis. Science 300, 486–489 (2003).
Paramithiotis, E. et al. A prion protein epitope selective for the pathologically misfolded conformation. Nat. Med. 9, 893–899 (2003).
Deng, H.X. et al. Amyotrophic lateral sclerosis and structural defects in Cu,Zn superoxide dismutase. Science 261, 1047–1051 (1993).
Gurney, M.E. et al. Motor neuron degeneration in mice that express a human Cu,Zn superoxide dismutase mutation. Science 264, 1772–1775 (1994).
Wong, P.C. et al. An adverse property of a familial ALS-linked SOD1 mutation causes motor neuron disease characterized by vacuolar degeneration of mitochondria. Neuron 14, 1105–1116 (1995).
Bruijn, L.I. et al. ALS-linked SOD1 mutant G85R mediates damage to astrocytes and promotes rapidly progressive disease with SOD1-containing inclusions. Neuron 18, 327–338 (1997).
Tiwari, A., Xu, Z. & Hayward, L.J. Aberrantly increased hydrophobicity shared by mutants of Cu,Zn-superoxide dismutase in familial amyotrophic lateral sclerosis. J. Biol. Chem. 280, 29771–29779 (2005).
Jonsson, P.A. et al. Minute quantities of misfolded mutant superoxide dismutase-1 cause amyotrophic lateral sclerosis. Brain 127, 73–88 (2004).
Furukawa, Y., Torres, A.S. & O'Halloran, T.V. Oxygen-induced maturation of SOD1: a key role for disulfide formation by the copper chaperone CCS. EMBO J. 23, 2872–2881 (2004).
Jaarsma, D. et al. Human Cu/Zn superoxide dismutase (SOD1) overexpression in mice causes mitochondrial vacuolization, axonal degeneration, and premature motoneuron death and accelerates motoneuron disease in mice expressing a familial amyotrophic lateral sclerosis mutant SOD1. Neurobiol. Dis. 7, 623–643 (2000).
Deng, H.X. et al. Conversion to the amyotrophic lateral sclerosis phenotype is associated with intermolecular linked insoluble aggregates of SOD1 in mitochondria. Proc. Natl. Acad. Sci. USA 103, 7142–7147 (2006).
Higgins, C.M., Jung, C. & Xu, Z. ALS-associated mutant SOD1G93A causes mitochondrial vacuolation by expansion of the intermembrane space and by involvement of SOD1 aggregation and peroxisomes. BMC Neurosci. 4, 16 (2003).
Bergemalm, D. et al. Overloading of stable and exclusion of unstable human superoxide dismutase-1 variants in mitochondria of murine amyotrophic lateral sclerosis models. J. Neurosci. 26, 4147–4154 (2006).
Kikuchi, H. et al. Spinal cord endoplasmic reticulum stress associated with a microsomal accumulation of mutant superoxide dismutase-1 in an ALS model. Proc. Natl. Acad. Sci. USA 103, 6025–6030 (2006).
Atkin, J.D. et al. Induction of the unfolded protein response in familial amyotrophic lateral sclerosis and association of protein disulfide isomerase with superoxide dismutase 1. J. Biol. Chem. 281, 30152–30165 (2006).
Boillee, S. et al. Onset and progression in inherited ALS determined by motor neurons and microglia. Science 312, 1389–1392 (2006).
Rudiger, S., Germeroth, L., Schneider-Mergener, J. & Bukau, B. Substrate specificity of the DnaK chaperone determined by screening cellulose-bound peptide libraries. EMBO J. 16, 1501–1507 (1997).
Acknowledgements
We thank M. Strong (University of Western Ontario) for providing human spinal cord sections and P.H. Chan (Stanford University) for providing human wild-type SOD1 rats. We also thank N. Ng and members of the Chakrabartty lab and the Centre for Research in Neurodegenerative Diseases for helpful comments and review; V. Mulligan, J. Kim and W. Zhou for technical assistance; and C. Accardi for assistance with animal care. This project was funded by the Neuromuscular Research Partnership—the Canadian Institutes of Health Research (CIHR), the ALS Society (Canada) and the Muscular Dystrophy Association (Canada) (J.R. and A.C.); the ALS Association (US) and MND Association (UK) (J.R.); and CIHR and the Temerety Family Trust (N.R.C). C.V.V. is funded by the Muscular Dystrophy Association and D.W.C. receives salary support from the Ludwig Institute. R.R. is the recipient of a Doctoral Research Award from the CIHR Institute of Neuroscience, Mental Health and Addiction (CIHR-INMHA) and the ALS Society (Canada). J.R. holds a Canada Research Chair in Molecular Mechanisms of ALS. N.R.C. holds a Canada Research Chair in Neurodegeneration and Protein Misfolding Diseases.
Author information
Authors and Affiliations
Contributions
R.R., J.R., D.W.C., N.R.C. and A.C. designed the research. R.R., J.R., C.V.V., P.H., D.M.R. and J.G. performed the research. R.R., J.R., C.V.V., D.W.C., N.R.C. and A.C. analyzed the data. R.R., J.R., C.V.V., D.W.C., N.R.C. and A.C. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
R.R., N.R.C. and A.C. are named as inventors on patent applications relating to this technology. The University Health Network has licensed this patent to a private-sector company that provides research funding; however, no funding from this company was used in executing the work described in this paper.
Supplementary information
Supplementary Fig. 1
SOD1 unfolding was followed by changes to tryptophan fluorescence (ex. = 280nm, em. = 350nm). (PDF 640 kb)
Supplementary Fig. 2
Western blots were performed to further demonstrate the specificity of the SEDI antibody. (PDF 1011 kb)
Supplementary Fig. 3
Comparison of immunoprecipitation reactions using SEDI antibody or pre-immune IgG. (PDF 937 kb)
Supplementary Fig. 4
SEDI antibody immunohistological labeling is specific and can be saturated by competition with the antigenic peptide. (PDF 1516 kb)
Supplementary Fig. 5
SEDI labeling of a mutant SOD1-transgenic mouse spinal cords. (PDF 3472 kb)
Supplementary Fig. 6
Our immunohistochemistry experiments show that monomer/misfolded SOD1 accumulates around vacuoles (Fig. 3b). (PDF 1420 kb)
Supplementary Fig. 7
Putative Hsp70 binding sites in SOD1 – 4-5 hydrophobic residues flanked by basic residues. (PDF 961 kb)
Rights and permissions
About this article
Cite this article
Rakhit, R., Robertson, J., Velde, C. et al. An immunological epitope selective for pathological monomer-misfolded SOD1 in ALS. Nat Med 13, 754–759 (2007). https://doi.org/10.1038/nm1559
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nm1559
This article is cited by
-
Selective removal of misfolded SOD1 delays disease onset in a mouse model of amyotrophic lateral sclerosis
Cellular and Molecular Life Sciences (2023)
-
Wild-type Cu/Zn-superoxide dismutase is misfolded in cerebrospinal fluid of sporadic amyotrophic lateral sclerosis
Molecular Neurodegeneration (2019)
-
Cu/Zn-superoxide dismutase and wild-type like fALS SOD1 mutants produce cytotoxic quantities of H2O2 via cysteine-dependent redox short-circuit
Scientific Reports (2019)
-
Impaired Cu–Zn Superoxide Dismutase (SOD1) and Calcineurin (Cn) Interaction in ALS: A Presumed Consequence for TDP-43 and Zinc Aggregation in Tg SOD1G93A Rodent Spinal Cord Tissue
Neurochemical Research (2019)
-
Misfolded SOD1 pathology in sporadic Amyotrophic Lateral Sclerosis
Scientific Reports (2018)