Abstract
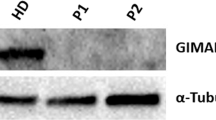
Lysosome-related organelles have versatile functions, including protein and lipid degradation, signal transduction and protein secretion. The molecular elucidation of rare congenital diseases affecting endosomal-lysosomal biogenesis has given insights into physiological functions of the innate and adaptive immune system. Here, we describe a previously unknown human primary immunodeficiency disorder and provide evidence that the endosomal adaptor protein p14, previously characterized as confining mitogen-activated protein kinase (MAPK) signaling to late endosomes, is crucial for the function of neutrophils, B cells, cytotoxic T cells and melanocytes. Combining genetic linkage studies and transcriptional profiling analysis, we identified a homozygous point mutation in the 3′ untranslated region (UTR) of p14 (also known as MAPBPIP), resulting in decreased protein expression. In p14-deficient cells, the distribution of late endosomes was severely perturbed, suggesting a previously unknown role for p14 in endosomal biogenesis. These findings have implications for understanding endosomal membrane dynamics, compartmentalization of cell signal cascades, and their role in immunity.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Accession codes
References
Fischer, A. Human primary immunodeficiency diseases: a perspective. Nat. Immunol. 5, 23–30 (2004).
Stinchcombe, J., Bossi, G. & Griffiths, G.M. Linking albinism and immunity: the secrets of secretory lysosomes. Science 305, 55–59 (2004).
Barbosa, M.D.F.S. et al. Identification of the homologous beige and Chediak-Higashi syndrome genes. Nature 382, 262–265 (1996).
Nagle, D.L. et al. Identification and mutation analysis of the complete gene for Chediak-Higashi syndrome. Nat. Genet. 14, 307–311 (1996).
Klein, C. et al. Partial albinism with immunodeficiency (Griscelli syndrome). J. Pediatr. 125, 886–895 (1994).
Ménasché, G. et al. Mutations in RAB27A cause Griscelli syndrome associated with haemophagocytic syndrome. Nat. Genet. 25, 173–176 (2000).
Dell'Angelica, E.C., Shotelersuk, V., Aguilar, R.C., Gahl, W.A. & Bonifacino, J.S. Altered trafficking of lysosomal proteins in Hermansky-Pudlak syndrome due to mutations in the β3A subunit of the AP-3 adaptor. Mol. Cell 3, 11–21 (1999).
Jung, J. et al. Identification of a homozygous deletion in the AP3B1 gene causing Hermansky-Pudlak syndrome, type 2. Blood 108, 362–369 (2006).
Kornfeld, S. & Mellman, I. The biogenesis of lysosomes. Annu. Rev. Cell Biol. 5, 483–525 (1989).
Janvier, K. & Bonifacino, J.S. Role of the endocytic machinery in the sorting of lysosome-associated membrane proteins. Mol. Biol. Cell 16, 4231–4242 (2005).
Bonifacino, J.S. & Glick, B.S. The mechanisms of vesicle budding and fusion. Cell 116, 153–166 (2004).
Ghosh, P., Dahms, N.M. & Kornfeld, S. Mannose 6-phosphate receptors: new twists in the tale. Nat. Rev. Mol. Cell Biol. 4, 202–212 (2003).
Gruenberg, J. & Stenmark, H. The biogenesis of multivesicular endosomes. Nat. Rev. Mol. Cell Biol. 5, 317–323 (2004).
Mor, A. & Philips, M.R. Compartmentalized Ras/MAPK signalling. Annu. Rev. Immunol. 24, 771–800 (2006).
Kolch, W. Coordinating ERK/MAPK signalling through scaffolds and inhibitors. Nat. Rev. Mol. Cell Biol. 6, 827–837 (2005).
Wunderlich, W. et al. A novel 14-kilodalton protein interacts with the mitogen-activated protein kinase scaffold mp1 on a late endosomal/lysosomal compartment. J. Cell Biol. 152, 765–776 (2001).
Teis, D., Wunderlich, W. & Huber, L.A. Localization of the MP1-MAPK scaffold complex to endosomes is mediated by p14 and required for signal transduction. Dev. Cell 3, 803–814 (2002).
Kurzbauer, R. et al. Crystal structure of the p14/MP1 scaffolding complex: how a twin couple attaches mitogen-activated protein kinase signaling to late endosomes. Proc. Natl. Acad. Sci. USA 101, 10984–10989 (2004).
Russell, J.H. & Ley, T.J. Lymphocyte-mediated cytotoxicity. Annu. Rev. Immunol. 20, 323–370 (2002).
Trambas, C.M. & Griffiths, G.M. Delivering the kiss of death. Nat. Immunol. 4, 399–403 (2003).
Hamers, M.N., Bot, A.A., Weening, R.S., Sips, H.J. & Roos, D. Kinetics and mechanism of the bactericidal action of human neutrophils against Escherichia coli. Blood 64, 635–641 (1984).
Chen, J.-M., Férec, C. & Cooper, D.N. A systematic analysis of disease-associated variants in the 3′ regulatory regions of human protein-coding genes I: general principles and overview. Hum. Genet. 120, 1–21 (2006).
Souza, L.M. et al. Recombinant human granulocyte colony-stimulating factor: effects on normal and leukemic myeloid cells. Science 232, 61–65 (1986).
Semerad, C.L., Liu, F., Gregory, A.D., Stumpf, K. & Link, D.C. G-CSF is an essential regulator of neutrophil trafficking from the bone marrow to the blood. Immunity 17, 413–423 (2002).
Aarts, L.H.J., Roovers, O., Ward, A.C. & Touw, I.P. Receptor activation and 2 distinct COOH-terminal motifs control G-CSF receptor distribution and internalization kinetics. Blood 103, 571–579 (2004).
Teis, D. et al. p14-MP1-MEK1 signaling regulates endosomal traffic and cellular proliferation during tissue homeostasis. J. Cell Biol. (in the press).
Price, T.H., Chatta, G.S. & Dale, D.C. Effect of recombinant granulocyte colony-stimulating factor on neutrophil kinetics in normal young and elderly humans. Blood 88, 335–340 (1996).
Carlsson, G. et al. Kostmann syndrome: severe congenital neutropenia associated with defective expression of Bcl-2, constitutive mitochondrial release of cytochrome c, and excessive apoptosis of myeloid progenitor cells. Blood 103, 3355–3361 (2004).
Klein, C. et al. Deficiency of HAX1 causes severe congenital neutropenia (Kostmann disease). Nat. Genet. (in the press).
Köllner, I. et al. Mutations in neutrophil elastase causing congenital neutropenia lead to cytoplasmic protein accumulation and induction of the unfolded protein response. Blood 108, 493–500 (2006).
Zhuang, D., Qiu, Y., Kogan, S.C. & Dong, F. Increased CCAAT enhancer-binding protein epsilon (C/EBPepsilon) expression and premature apoptosis in myeloid cells expressing Gfi-1 N382S mutant associated with severe congenital neutropenia. J. Biol. Chem. 281, 10745–10751 (2006).
Huynh, C., Roth, D., Ward, D.M., Kaplan, J. & Andrews, N.W. Defective lysosomal exocytosis and plasma membrane repair in Chediak-Higashi/beige cells. Proc. Natl. Acad. Sci. USA 101, 16795–16800 (2004).
Baetz, K., Isaatz, S. & Griffiths, G.M. Loss of cytotoxic T lymphocyte function in Chediak Higashi syndrome arises from a secretory defect that prevents lytic granule exocytosis. J. Immunol. 154, 6122–6131 (1995).
Haddad, E.K., Wu, X., Hammer, J.A., III. & Henkart, P.A. Defective granule exocytosis in Rab27a-deficient lymphocytes from ashen mice. J. Cell Biol. 152, 835–842 (2001).
Stinchcombe, J. et al. Rab27a is required for regulated secretion in cytotoxic T lymphocytes. J. Cell Biol. 152, 825–834 (2001).
O'Connell, J.R. & Weeks, D.E. PedCheck: A program for identification of genotype incompatibilities in linkage analysis. Am. J. Hum. Genet. 63, 259–266 (1998).
Lathrop, G.M. & Lalouel, J.-M. Easy calculations of LOD scores and genetic risks on small computers. Am. J. Hum. Genet. 36, 460–465 (1984).
Cottingham, R.W., Idury, R.M., Jr. & Schäffer, A.A. Faster sequential genetic linkage computations. Am. J. Hum. Genet. 53, 252–263 (1993).
Schäffer, A.A., Gupta, S.K., Shriram, K. & Cottingham, R.W., Jr. Avoiding recomputation in linkage analysis. Hum. Hered. 44, 225–237 (1994).
Hino, M. et al. Ex vivo expansion of mature human neutrophils with normal functions from purified peripheral blood CD34+ haematopoietic progenitor cells. Br. J. Haematol. 109, 314–321 (2000).
Klein, C., Bueeler, H. & Mulligan, R.C. Comparative analysis of genetically modified dendritic cells and cytokine transduced tumor cells as therapeutic cancer vaccines. J. Exp. Med. 191, 1699–1708 (2000).
Ory, D.S., Neugeboren, B.A. & Mulligan, R.C. A stable human-derived packaging cell line for production of high titer retrovirus/vesicular stomatitis virus G pseudotypes. Proc. Natl. Acad. Sci. USA 93, 11400–11406 (1996).
Acknowledgements
We thank M. Schlesier (University of Freiburg) for the B-cell phenotyping, I. Sandrock, G. Köhne, F. Noyan, K. Boztug and M. Ballmaier for laboratory support, C. Roifman (Hospital for Sick Children) and R. Gatti (University of California at Los Angeles) for providing control samples on Mennonite families, and E. Ungewickell, C. Baum, J. Bohne, C. Kardinal and H. Holtmann for critical discussions. We thank B. Tümmler (Hannover Medical School), C. Baum (Hannover Medical School) and I. Touw (Erasmus University Medical Center) for reagents, M. Zimmermann for help in statistical evaluations and M. Offterdinger for help in determining subcellular distribution of endosomes. This research was supported by Deutsche Forschungsgemeinschaft grants KFO110 and GR1617/3, BMBF, the Elternverein Krebskranke Kinder Hannover, the Austrian Proteomics Platform (APP, GEN-AU), the Special Research Program “Cell Proliferation and Cell Death in Tumors” (SFB021, Austrian Science Fund) and in part by the intramural research program of the US National Institutes of Health, National Library of Medicine (NLM).
Author information
Authors and Affiliations
Contributions
A.A., G. Brandes and J.T. contributed equally to this work. G. Bohn sequenced candidate genes and performed most molecular and cellular functional studies. A.A. characterized the 3′ UTR mutation by assessing RNA metabolism and functional reporter assays. G. Brandes performed electron microscopy and immunofluorescence studies. J.T. performed fine-mapping, screened candidate genes and helped to edit the manuscript. E.G. performed the genome-wide scan. A.A.S. carried out the genetic linkage analysis computations, chose the markers for genetic linkage fine mapping and wrote parts of the manuscript. C.R. performed E. coli lysis assays. N.T. and D.T. did immunofluorescence studies on endosomes. C.Z. cared for patients, and collected and curated data in SCN patient registry. R.A.D. assisted Bohn, A.A. and C.R. R.G. and J.B. performed microarray experiments. L.A.H. gave advice on endosome biology, and educated and supervised N.T. and D.T. K.W. provided laboratory resources, resources for SCN registry and significant help to initiate and carry out the study. B.G. initiated the project together with C.K., educated and supervised J.T. and E.G., assisted A.A.S. in linkage analysis, provided grant and laboratory resources and edited the manuscript. C.K. designed and directed the study, obtained clinical samples, taught and supervised Bohn, A.A., C.R. and R.A.D., provided laboratory and financial resources and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Fig. 1
Assessment of granzyme B and perforin in cytotoxic T-cells, and TEM of neutrophil granulocytes of patients #10, 12 and 13. (PDF 1673 kb)
Supplementary Fig. 2
Statistical analysis of E. coli lysis assay. Data presented in Fig. 4b were transformed by grouping the samples from HD and genetically corrected cells from patient #5 (black lines). (PDF 61 kb)
Supplementary Fig. 3
Analysis of G-CSF-receptor (G-CSFR) transgenic fibroblast cell lines. (PDF 88 kb)
Supplementary Figure 4
Statistical analysis of endosome distribution with respect to the distance from the nucleus. (PDF 2523 kb)
Supplementary Table 1
Immunophenotyping, in vitro T-cell proliferation, serum immunoglobulin levels and vaccination titers. (PDF 116 kb)
Supplementary Table 2
LOD scores and target genes. (PDF 98 kb)
Supplementary Table 3
Transcriptional profiling of EBV-transformed B-cell lines. Expression intensities of transcripts in EBV-transformed B-cell lines derived from parents (#1, 2) werecompared to B-cell lines derived from two patients (#12, 13). (PDF 131 kb)
Rights and permissions
About this article
Cite this article
Bohn, G., Allroth, A., Brandes, G. et al. A novel human primary immunodeficiency syndrome caused by deficiency of the endosomal adaptor protein p14. Nat Med 13, 38–45 (2007). https://doi.org/10.1038/nm1528
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nm1528
This article is cited by
-
The Ragulator complex: delving its multifunctional impact on metabolism and beyond
Inflammation and Regeneration (2023)
-
The lysosomal Ragulator complex plays an essential role in leukocyte trafficking by activating myosin II
Nature Communications (2021)
-
Limited survival and impaired hepatic fasting metabolism in mice with constitutive Rag GTPase signaling
Nature Communications (2021)
-
Translation deregulation in human disease
Nature Reviews Molecular Cell Biology (2018)