Abstract
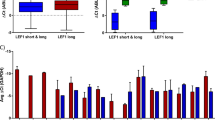
Interstitial loss of all or part of the long arm of chromosome 5, or del(5q), is a frequent clonal chromosomal abnormality in human myelodysplastic syndrome (MDS, a preleukemic disorder) and acute myeloid leukemia (AML)1, and is thought to contribute to the pathogenesis of these diseases by deleting one or more tumor-suppressor genes2. Although a major commonly deleted region (CDR) has been delineated on chromosome band 5q31.1 (refs. 3–7), attempts to identify tumor suppressors within this band have been unsuccessful. We focused our analysis of gene expression on RNA from primitive leukemia-initiating cells, which harbor 5q deletions8,9, and analyzed 12 genes within the CDR that are expressed by normal hematopoietic stem cells. Here we show that the gene encoding α-catenin (CTNNA1) is expressed at a much lower level in leukemia-initiating stem cells from individuals with AML or MDS with a 5q deletion than in individuals with MDS or AML lacking a 5q deletion or in normal hematopoietic stem cells. Analysis of HL-60 cells, a myeloid leukemia line with deletion of the 5q31 region10,11, showed that the CTNNA1 promoter of the retained allele is suppressed by both methylation and histone deacetylation. Restoration of CTNNA1 expression in HL-60 cells resulted in reduced proliferation and apoptotic cell death. Thus, loss of expression of the α-catenin tumor suppressor in hematopoietic stem cells may provide a growth advantage that contributes to human MDS or AML with del(5q).
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Nimer, S.D. & Golde, D.W. The 5q- abnormality. Blood 70, 1705–1712 (1987).
Van den Berghe, H. & Michaux, L. 5q-, twenty-five years later: a synopsis. Cancer Genet. Cytogenet. 94, 1–7 (1997).
Le Beau, M.M. et al. Cytogenetic and molecular delineation of the smallest commonly deleted region of chromosome 5 in malignant myeloid diseases. Proc. Natl. Acad. Sci. USA 90, 5484–5488 (1993).
Horrigan, S.K. et al. Delineation of a minimal interval and identification of 9 candidates for a tumor suppressor gene in malignant myeloid disorders on 5q31. Blood 95, 2372–2377 (2000).
Zhao, N. et al. Molecular delineation of the smallest commonly deleted region of chromosome 5 in malignant myeloid diseases to 1–1.5 Mb and preparation of a PAC-based physical map. Proc. Natl. Acad. Sci. USA 94, 6948–6953 (1997).
Fairman, J., Chumakov, I., Chinault, A.C., Nowell, P.C. & Nagarajan, L. Physical mapping of the minimal region of loss in 5q- chromosome. Proc. Natl. Acad. Sci. USA 92, 7406–7410 (1995).
Boultwood, J. et al. Narrowing and genomic annotation of the commonly deleted region of the 5q- syndrome. Blood 99, 4638–4641 (2002).
Bonnet, D. & Dick, J.E. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat. Med. 3, 730–737 (1997).
Nilsson, L. et al. Isolation and characterization of hematopoietic progenitor/stem cells in 5q-deleted myelodysplastic syndromes: evidence for involvement at the hematopoietic stem cell level. Blood 96, 2012–2021 (2000).
Shipley, J., Weber-Hall, S. & Birdsall, S. Loss of the chromosomal region 5q11-q31 in the myeloid cell line HL-60: characterization by comparative genomic hybridization and fluorescence in situ hybridization. Genes Chromosom. Cancer 15, 182–186 (1996).
Liang, J.C. et al. Spectral karyotypic study of the HL-60 cell line: detection of complex rearrangements involving chromosomes 5, 7, and 16 and delineation of critical region of deletion on 5q31.1. Cancer Genet. Cytogenet. 113, 105–109 (1999).
Lapidot, T. et al. A cell initiating human acute myeloid leukaemia after transplantation into SCID mice. Nature 367, 645–648 (1994).
Blair, A., Hogge, D.E., Ailles, L.E., Lansdorp, P.M. & Sutherland, H.J. Lack of expression of Thy-1 (CD90) on acute myeloid leukemia cells with long-term proliferative ability in vitro and in vivo. Blood 89, 3104–3112 (1997).
Jordan, C.T. et al. The interleukin-3 receptor alpha chain is a unique marker for human acute myelogenous leukemia stem cells. Leukemia 14, 1777–1784 (2000).
Mrózek, K., Tanner, S.M., Heinonen, K. & Bloomfield, C.D. Molecular cytogenetic characterization of the KG-1 and KG-1a acute myeloid leukemia cell lines by use of spectral karyotyping and fluorescence in situ hybridization. Genes Chromosom. Cancer 38, 249–252 (2003).
Dick, J.E. Stem cells: Self-renewal writ in blood. Nature 423, 231–233 (2003).
Lai, F. et al. Transcript map and comparative analysis of the 1.5-Mb commonly deleted segment of human 5q31 in malignant myeloid diseases with a del(5q). Genomics 71, 235–245 (2001).
Ulger, C. et al. Comprehensive genome-wide comparison of DNA and RNA level scan using microarray technology for identification of candidate cancer-related genes in the HL-60 cell line. Cancer Genet. Cytogenet. 147, 28–35 (2003).
Lee, J.Y. et al. Molecular cytogenetic analysis of the monoblastic cell line U937. karyotype clarification by G-banding, whole chromosome painting, microdissection and reverse painting, and comparative genomic hybridization. Cancer Genet. Cytogenet. 137, 124–132 (2002).
Vanpoucke, G., Nollet, F., Tejpar, S., Cassiman, J.J. & van Roy, F. The human alphaE-catenin gene CTNNA1: mutational analysis and rare occurrence of a truncated splice variant. Biochim. Biophys. Acta 1574, 262–268 (2002).
Lu, B., Roegiers, F., Jan, L.Y. & Jan, Y.N. Adherens junctions inhibit asymmetric division in the Drosophila epithelium. Nature 409, 522–525 (2001).
Yamashita, Y.M., Jones, D.L. & Fuller, M.T. Orientation of asymmetric stem cell division by the APC tumor suppressor and centrosome. Science 301, 1547–1550 (2003).
Vasioukhin, V., Bauer, C., Degenstein, L., Wise, B. & Fuchs, E. Hyperproliferation and defects in epithelial polarity upon conditional ablation of alpha-catenin in skin. Cell 104, 605–617 (2001).
Lechler, T. & Fuchs, E. Asymmetric cell divisions promote stratification and differentiation of mammalian skin. Nature 437, 275–280 (2005).
Zhang, J. et al. Identification of the haematopoietic stem cell niche and control of the niche size. Nature 425, 836–841 (2003).
Clark, S.J., Harrison, J., Paul, C.L. & Frommer, M. High sensitivity mapping of methylated cytosines. Nucleic Acids Res. 22, 2990–2997 (1994).
Shu, J. et al. Silencing of bidirectional promoters by DNA methylation in tumorigenesis. Cancer Res. 66, 5077–5084 (2006).
Acknowledgements
We are grateful to S. Heinrichs and K. Griffith for statistical analysis; D. Adams and the University of Michigan flow cytometry core for assistance in the design and implementation of the sort paradigms used in this study; K. Mrózek for cytogenetic review; L. Moreau and J. Eskenas for assistance with fluorescence in situ hybridization; and J. Gilbert and J. Berman for editorial assistance and critical comments. This work was supported by a Leukemia and Lymphoma Society SCOR grant, the National Natural Science Foundation of China (30525019 to T.X.L.), the Leukemia Clinical Research Foundation and the US National Institutes of Health (grants CA108631 to A.T.L. and J.-P.I.; CA104987 to M.F.C.; and CA101140 to C.D.B.).
Author information
Authors and Affiliations
Contributions
T.X.L. conducted the RT-PCR, functional experiments and wrote the manuscript. M.W.B. performed the hematopoietic cell sorting and real-time PCR, and wrote the manuscript. J.J. and J.-P.I. did the methylation analysis. W.-S.W. performed the retroviral experiments. M.D. helped with the RT-PCR experiments. M.F.C. and A.T.L. supervised the project. Others contributed to the handling of patient samples and to the editing of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Fig. 1
Expression profile of 28 5q CDR genes in L-ICs derived from del(5q)-V. (PDF 1063 kb)
Supplementary Fig. 2
Immunofluorescence assay of α-catenin protein in sorted normal HSC, and L-ICs. (PDF 1221 kb)
Supplementary Fig. 3
Level of CTNNA1 expression in normal HSCs (CD34+ CD38− Lin−) and leukemia cell lines either untreated or treated with 0.5 μM DAC for 48 hr or 0.3 μM TSA for 24 hr. (PDF 177 kb)
Supplementary Fig. 4
Model for the malignant transformation of α-catenin-deficient HSCs. (PDF 900 kb)
Supplementary Table 1
Clinical characteristics of MDS/AML patients with or without del(5q) (PDF 132 kb)
Supplementary Table 2
Frequency of 5q deletion in HL-60 cells and AML/MDS patients as determined by D-1 FISH. (PDF 95 kb)
Supplementary Table 3
Primer sequences of genes located in 5q31.1 commonly deleted region. (PDF 99 kb)
Rights and permissions
About this article
Cite this article
Liu, T., Becker, M., Jelinek, J. et al. Chromosome 5q deletion and epigenetic suppression of the gene encoding α-catenin (CTNNA1) in myeloid cell transformation. Nat Med 13, 78–83 (2007). https://doi.org/10.1038/nm1512
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nm1512
This article is cited by
-
DELE1 haploinsufficiency causes resistance to mitochondrial stress-induced apoptosis in monosomy 5/del(5q) AML
Leukemia (2023)
-
Decitabine combined with minimally myelosuppressive therapy for induction of remission in pediatric high-risk acute myeloid leukemia with chromosome 5q deletion: a report of three cases
International Journal of Hematology (2022)
-
Genome-wide DNA methylation analysis pre- and post-lenalidomide treatment in patients with myelodysplastic syndrome with isolated deletion (5q)
Annals of Hematology (2021)
-
Age-related inflammatory bone marrow microenvironment induces ineffective erythropoiesis mimicking del(5q) MDS
Leukemia (2018)
-
α-catenin SUMOylation increases IκBα stability and inhibits breast cancer progression
Oncogenesis (2018)