Abstract
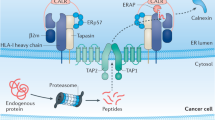
Defects in major histocompatibility complex (MHC) class I–restricted antigen presentation are frequently observed in human cancers and result in escape of tumors from cytotoxic T lymphocyte (CTL) immune surveillance in mice. Here, we show the existence of a unique category of CTLs that can prevent this escape. The CTLs target an alternative repertoire of peptide epitopes that emerge in MHC class I at the surface of cells with impaired function of transporter associated with antigen processing (TAP), tapasin or the proteasome. These peptides, although derived from self antigens such as the commonly expressed Lass5 protein (also known as Trh4), are not presented by normal cells. This explains why they act as immunogenic neoantigens. The newly discovered epitopes can be exploited for immune intervention against processing-deficient tumors through adoptive T-cell transfer or peptide vaccination.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Accession codes
References
Melief, C.J. et al. Strategies for immunotherapy of cancer. Adv. Immunol. 75, 235–282 (2000).
Khong, H.T. & Restifo, N.P. Natural selection of tumor variants in the generation of “tumor escape” phenotypes. Nat. Immunol. 3, 999–1005 (2002).
Hicklin, D.J., Marincola, F.M. & Ferrone, S. HLA class I antigen downregulation in human cancers: T-cell immunotherapy revives an old story. Mol. Med. Today 5, 178–186 (1999).
Seliger, B., Maeurer, M.J. & Ferrone, S. TAP off–tumors on. Immunol. Today 18, 292–299 (1997).
Marincola, F.M., Jaffee, E.M., Hicklin, D.J. & Ferrone, S. Escape of human solid tumors from T-cell recognition: molecular mechanisms and functional significance. Adv. Immunol. 74, 181–273 (2000).
Cresswell, P., Bangia, N., Dick, T. & Diedrich, G. The nature of the MHC class I peptide loading complex. Immunol. Rev. 172, 21–28 (1999).
Van Kaer, L., Ashton-Rickardt, P.G., Ploegh, H.L. & Tonegawa, S. TAP1 mutant mice are deficient in antigen presentation, surface class I molecules, and CD4−8+ T cells. Cell 71, 1205–1214 (1992).
Chen, H.L. et al. A functionally defective allele of TAP1 results in loss of MHC class I antigen presentation in a human lung cancer. Nat. Genet. 13, 210–213 (1996).
Evans, M. et al. Antigen processing defects in cervical carcinomas limit the presentation of a CTL epitope from human papillomavirus 16 E6. J. Immunol. 167, 5420–5428 (2001).
Alimonti, J. et al. TAP expression provides a general method for improving the recognition of malignant cells in vivo. Nat. Biotechnol. 18, 515–520 (2000).
Wolpert, E.Z. et al. Generation of CD8+ T cells specific for transporter associated with antigen processing deficient cells. Proc. Natl. Acad. Sci. USA 94, 11496–11501 (1997).
Vivier, E. & Anfossi, N. Inhibitory NK-cell receptors on T cells: witness of the past, actors of the future. Nat. Rev. Immunol. 4, 190–198 (2004).
Reits, E.A., Vos, J.C., Gromme, M. & Neefjes, J. The major substrates for TAP in vivo are derived from newly synthesized proteins. Nature 404, 774–778 (2000).
Garbi, N. et al. Impaired immune responses and altered peptide repertoire in tapasin-deficient mice. Nat. Immunol. 1, 234–238 (2000).
Howell, D. et al. Heterogeneity of RMA-S cell line: derivatives of RMA-S cells lacking H2-Kb and H2-Db expression. Immunogenetics 52, 150–154 (2000).
Chiang, E.Y., Henson, M. & Stroynowski, I. Correction of defects responsible for impaired Qa-2 class Ib MHC expression on melanoma cells protects mice from tumor growth. J. Immunol. 170, 4515–4523 (2003).
Winter, E. & Ponting, C.P. TRAM, LAG1 and CLN8: members of a novel family of lipid-sensing domains? Trends Biochem. Sci. 27, 381–383 (2002).
Riebeling, C., Allegood, J.C., Wang, E., Merrill, A.H., Jr. & Futerman, A.H. Two mammalian longevity assurance gene (LAG1) family members, trh1 and trh4, regulate dihydroceramide synthesis using different fatty acyl-CoA donors. J. Biol. Chem. 278, 43452–43459 (2003).
Snyder, H.L., Bacik, I., Yewdell, J.W., Behrens, T.W. & Bennink, J.R. Promiscuous liberation of MHC-class I-binding peptides from the C termini of membrane and soluble proteins in the secretory pathway. Eur. J. Immunol. 28, 1339–1346 (1998).
Zwaveling, S. et al. Established human papillomavirus type 16-expressing tumors are effectively eradicated following vaccination with long peptides. J. Immunol. 169, 350–358 (2002).
Ossendorp, F., Mengede, E., Camps, M., Filius, R. & Melief, C.J. Specific T-helper cell requirement for optimal induction of cytotoxic T-lymphocytes against major histocompatibility complex class-II negative tumors. J. Exp. Med. 187, 693–702 (1998).
De Silva, A.D. et al. Thermolabile H-2Kb molecules expressed by transporter associated with antigen processing-deficient RMA-S cells are occupied by low-affinity peptides. J. Immunol. 163, 4413–4420 (1999).
Yewdell, J.W., Reits, E. & Neefjes, J.J. Making sense of mass destruction: quantitating MHC class I antigen presentation. Nat. Rev. Immunol. 3, 952–961 (2003).
Snyder, H.L. et al. Two novel routes of transporter associated with antigen processing (TAP)-independent major histocompatibility complex class I antigen processing. J. Exp. Med. 186, 1087–1098 (1997).
Jensen, P.E., Sullivan, B.A., Reed-Loisel, L.M. & Weber, D.A. Qa-1, a nonclassical class I histocompatibility molecule with roles in innate and adaptive immunity. Immunol. Res. 29, 81–92 (2004).
Wei, M.L. & Cresswell, P. HLA-A2 molecules in an antigen-processing mutant cell contain signal sequence-derived peptides. Nature 356, 443–446 (1992).
Henderson, R.A. et al. HLA-A2.1-associated peptides from a mutant cell line: a second pathway of antigen presentation. Science 255, 1264–1266 (1992).
Johnsen, A.K., Templeton, D.J., Sy, M.S. & Harding, C.V. Deficiency of transporter for antigen presentation (TAP) in tumor cells allows evasion of immune surveillance and increased tumorigenesis. J. Immunol. 163, 4224–4231 (1999).
Qin, Z. et al. Increased tumorigenicity, but unchanged immunogenicity, of transporter for antigen presentation 1-deficient tumors. Cancer Res. 62, 2856–2860 (2002).
Van Hall, T. et al. Identification of a novel tumor-specific CTL epitope presented by RMA, EL-4, and MBL-2 lymhomas reveals their common origin. J. Immunol. 165, 869–877 (2000).
Glas, R., Sturmhofel, K., Hammerling, G.J., Karre, K. & Ljunggren, H.G. Restoration of a tumorigenic phenotype by beta 2-microglobulin transfection to EL-4 mutant cells. J. Exp. Med. 175, 843–846 (1992).
Van Hall, T. et al. Differential influence on cytotoxic T-lymphocyte epitope presentation by controlled expression of either proteasome immunosubunits or PA28. J. Exp. Med. 192, 483–494 (2000).
Summerton, J. & Weller, D. Morpholino antisense oligomers: design, preparation, and properties. Antisense Nucleic Acid Drug Dev. 7, 187–195 (1997).
Toomey, J.A. et al. Stochastic acquisition of Qa1 receptors during the development of fetal NK cells in vitro accounts in part but not in whole for the ability of these cells to distinguish between class I-sufficient and class I-deficient targets. J. Immunol. 163, 3176–3184 (1999).
Falk, K., Rotzschke, O., Stvanovic, S., Jung, G. & Rammensee, H.G. Allele-specific motifs revealed by sequencing of self-peptides eluted from MHC molecules. Nature 351, 290–296 (1991).
Meiring, H.D., Van der Heeft, E., Ten Hove, G.J. & De Jong, A.P. Nanoscale LC-MS: technical design and applications to peptide and protein analysis. J. Sep. Sci. 25, 557–568 (2002).
Hill, A. et al. Herpes simplex virus turns off the TAP to evade host immunity. Nature 375, 411–415 (1995).
Van Hall, T. et al. Cryptic open reading frames in plasmid vector backbone sequences can provide highly immunogenic cytotoxic T-lymphocyte epitopes. Cancer Res. 58, 3087–3093 (1998).
Acknowledgements
The authors would like to thank G.J. Hämmerling for providing knockout mice and S. Stevanovic for supply of motif-bearing peptide libraries. This work was financially supported by the Dutch Cancer Society (UL2002-2709 to T.v.H. and S.L.), the Centre for Medical Systems Biology (a centre of excellence approved by the Netherlands Genomics Initiative), the Swedish Cancer Foundation, the Swedish Medical Research Council, the Royal Swedish Academy of Sciences and Accuro Immunology AB.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Fig. 1
Kb- and Db-eluted peptides from TAP-deficient cells represent two different repertoires. (PDF 37 kb)
Supplementary Fig. 2
Mass spectrometry identification of three peptide sequences that are presented on TAP-deficient RMA-S cells. (PDF 208 kb)
Supplementary Fig. 3
The H-2Db-restricted TEIPP epitope is encoded by the Trh4 gene (PDF 116 kb)
Supplementary Fig. 4
Peptide MCLRMTAVM is detected in Db-eluted fractions from RMA-S. (PDF 220 kb)
Supplementary Table 1
Phenotypic characterization of TEIPP-specific CTL. (PDF 16 kb)
Supplementary Table 2
Identification of Kb-binding peptides from RMA-S. (PDF 11 kb)
Supplementary Table 3
Position screen in the mimotope peptide. (PDF 18 kb)
Supplementary Table 4
Determination of the Db peptide-epitope. (PDF 16 kb)
Rights and permissions
About this article
Cite this article
van Hall, T., Wolpert, E., van Veelen, P. et al. Selective cytotoxic T-lymphocyte targeting of tumor immune escape variants. Nat Med 12, 417–424 (2006). https://doi.org/10.1038/nm1381
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nm1381
This article is cited by
-
Vaccination against neoantigens induced in cross-priming cDC1 in vivo
Cancer Immunology, Immunotherapy (2024)
-
Multimodal immunogenomic biomarker analysis of tumors from pediatric patients enrolled to a phase 1-2 study of single-agent atezolizumab
Nature Cancer (2023)
-
Cross-presentation of a TAP-independent signal peptide induces CD8 T immunity to escaped cancers but necessitates anchor replacement
Cancer Immunology, Immunotherapy (2022)
-
HLA class I loss in colorectal cancer: implications for immune escape and immunotherapy
Cellular & Molecular Immunology (2021)
-
Tumor-targeted silencing of the peptide transporter TAP induces potent antitumor immunity
Nature Communications (2019)