Abstract
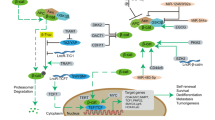
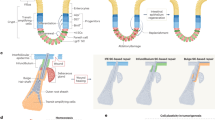
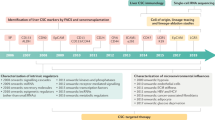
The cancer stem cell (CSC) concept was proposed four decades ago, and states that tumor growth, analogous to the renewal of healthy tissues, is fueled by small numbers of dedicated stem cells. It has gradually become clear that many tumors harbor CSCs in dedicated niches, and yet their identification and eradication has not been as obvious as was initially hoped. Recently developed lineage-tracing and cell-ablation strategies have provided insights into CSC plasticity, quiescence, renewal, and therapeutic response. Here we discuss new developments in the CSC field in relationship to changing insights into how normal stem cells maintain healthy tissues. Expectations in the field have become more realistic, and now, the first successes of therapies based on the CSC concept are emerging.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Clevers, H. The intestinal crypt, a prototype stem cell compartment. Cell 154, 274–284 (2013).
Doulatov, S., Notta, F., Laurenti, E. & Dick, J.E. Hematopoiesis: a human perspective. Cell Stem Cell 10, 120–136 (2012).
Uckun, F.M. et al. Leukemic cell growth in SCID mice as a predictor of relapse in high-risk B-lineage acute lymphoblastic leukemia. Blood 85, 873–878 (1995).
Lapidot, T. et al. A cell initiating human acute myeloid leukaemia after transplantation into SCID mice. Nature 367, 645–648 (1994).
Bonnet, D. & Dick, J.E. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat. Med. 3, 730–737 (1997).
Al-Hajj, M., Wicha, M.S., Benito-Hernandez, A., Morrison, S.J. & Clarke, M.F. Prospective identification of tumorigenic breast cancer cells. Proc. Natl. Acad. Sci. USA 100, 3983–3988 (2003).
O'Brien, C.A., Pollett, A., Gallinger, S. & Dick, J.E. A human colon cancer cell capable of initiating tumour growth in immunodeficient mice. Nature 445, 106–110 (2007).
Ricci-Vitiani, L. et al. Identification and expansion of human colon-cancer-initiating cells. Nature 445, 111–115 (2007).
Dalerba, P. et al. Phenotypic characterization of human colorectal cancer stem cells. Proc. Natl. Acad. Sci. USA 104, 10158–10163 (2007).
Singh, S.K. et al. Identification of human brain tumour initiating cells. Nature 432, 396–401 (2004).
Driessens, G., Beck, B., Caauwe, A., Simons, B.D. & Blanpain, C. Defining the mode of tumour growth by clonal analysis. Nature 488, 527–530 (2012).
Schepers, A.G. et al. Lineage tracing reveals Lgr5+ stem cell activity in mouse intestinal adenomas. Science 337, 730–735 (2012).
Kozar, S. et al. Continuous clonal labeling reveals small numbers of functional stem cells in intestinal crypts and adenomas. Cell Stem Cell 13, 626–633 (2013).
Zomer, A. et al. Intravital imaging of cancer stem cell plasticity in mammary tumors. Stem Cells 31, 602–606 (2013).
Chen, J. et al. A restricted cell population propagates glioblastoma growth after chemotherapy. Nature 488, 522–526 (2012).
Oshimori, N., Oristian, D. & Fuchs, E. TGF-b promotes heterogeneity and drug resistance in squamous cell carcinoma. Cell 160, 963–976 (2015).
Clevers, H. STEM CELLS. What is an adult stem cell? Science 350, 1319–1320 (2015).
Clevers, H. The cancer stem cell: premises, promises and challenges. Nat. Med. 17, 313–319 (2011).
Collins, A.T., Berry, P.A., Hyde, C., Stower, M.J. & Maitland, N.J. Prospective identification of tumorigenic prostate cancer stem cells. Cancer Res. 65, 10946–10951 (2005).
Li, C. et al. Identification of pancreatic cancer stem cells. Cancer Res. 67, 1030–1037 (2007).
Zhang, S. et al. Identification and characterization of ovarian cancer-initiating cells from primary human tumors. Cancer Res. 68, 4311–4320 (2008).
Eramo, A. et al. Identification and expansion of the tumorigenic lung cancer stem cell population. Cell Death Differ. 15, 504–514 (2008).
Nassar, D. & Blanpain, C. Cancer stem cells: basic concepts and therapeutic implications. Annu. Rev. Pathol. 11, 47–76 (2016).
Meacham, C.E. & Morrison, S.J. Tumour heterogeneity and cancer cell plasticity. Nature 501, 328–337 (2013).
Mascré, G. et al. Distinct contribution of stem and progenitor cells to epidermal maintenance. Nature 489, 257–262 (2012).
Clayton, E. et al. A single type of progenitor cell maintains normal epidermis. Nature 446, 185–189 (2007).
Barker, N. et al. Crypt stem cells as the cells-of-origin of intestinal cancer. Nature 457, 608–611 (2009).
Cortina, C. et al. A genome editing approach to study cancer stem cells in human tumors. EMBO Mol. Med. 9, 869–879 (2017).
Shimokawa, M. et al. Visualization and targeting of LGR5+ human colon cancer stem cells. Nature 545, 187–192 (2017).
Merlos-Suárez, A. et al. The intestinal stem cell signature identifies colorectal cancer stem cells and predicts disease relapse. Cell Stem Cell 8, 511–524 (2011).
Dalerba, P. et al. Single-cell dissection of transcriptional heterogeneity in human colon tumors. Nat. Biotechnol. 29, 1120–1127 (2011).
Barker, N., et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature 449, 1003–1007 (2007).
Barker, N. et al. Lgr5(+ve) stem cells drive self-renewal in the stomach and build long-lived gastric units in vitro. Cell Stem Cell 6, 25–36 (2010).
Doupé, D.P., Klein, A.M., Simons, B.D. & Jones, P.H. The ordered architecture of murine ear epidermis is maintained by progenitor cells with random fate. Dev. Cell 18, 317–323 (2010).
Leushacke, M., Ng, A., Galle, J., Loeffler, M. & Barker, N. Lgr5+ gastric stem cells divide symmetrically to effect epithelial homeostasis in the pylorus. Cell Reports 5, 349–356 (2013).
Snippert, H.J. et al. Intestinal crypt homeostasis results from neutral competition between symmetrically dividing Lgr5 stem cells. Cell 143, 134–144 (2010).
Lopez-Garcia, C., Klein, A.M., Simons, B.D. & Winton, D.J. Intestinal stem cell replacement follows a pattern of neutral drift. Science 330, 822–825 (2010).
Stanger, B.Z. Cellular homeostasis and repair in the mammalian liver. Annu. Rev. Physiol. 77, 179–200 (2015).
Brack, A.S. & Rando, T.A. Tissue-specific stem cells: lessons from the skeletal muscle satellite cell. Cell Stem Cell 10, 504–514 (2012).
Hsu, Y.C., Li, L. & Fuchs, E. Emerging interactions between skin stem cells and their niches. Nat. Med. 20, 847–856 (2014).
Tian, H. et al. A reserve stem cell population in small intestine renders Lgr5-positive cells dispensable. Nature 478, 255–259 (2011).
van Es, J.H. et al. Dll1+ secretory progenitor cells revert to stem cells upon crypt damage. Nat. Cell Biol. 14, 1099–1104 (2012).
Buczacki, S.J. et al. Intestinal label-retaining cells are secretory precursors expressing Lgr5. Nature 495, 65–69 (2013).
Tetteh, P.W. et al. Replacement of lost Lgr5-positive stem cells through plasticity of their enterocyte-lineage daughters. Cell Stem Cell 18, 203–213 (2016).
Sato, T. et al. Paneth cells constitute the niche for Lgr5 stem cells in intestinal crypts. Nature 469, 415–418 (2011).
Rock, J.R. et al. Basal cells as stem cells of the mouse trachea and human airway epithelium. Proc. Natl. Acad. Sci. USA 106, 12771–12775 (2009).
Tata, P.R. et al. Dedifferentiation of committed epithelial cells into stem cells in vivo. Nature 503, 218–223 (2013).
Kusaba, T., Lalli, M., Kramann, R., Kobayashi, A. & Humphreys, B.D. Differentiated kidney epithelial cells repair injured proximal tubule. Proc. Natl. Acad. Sci. USA 111, 1527–1532 (2014).
Quintana, E. et al. Efficient tumour formation by single human melanoma cells. Nature 456, 593–598 (2008).
Ball, C.R. et al. Succession of transiently active tumor-initiating cell clones in human pancreatic cancer xenografts. EMBO Mol. Med. 9, 918–932 (2017).
Gupta, P.B. et al. Stochastic state transitions give rise to phenotypic equilibrium in populations of cancer cells. Cell 146, 633–644 (2011).
van de Wetering, M. et al. The beta-catenin/TCF-4 complex imposes a crypt progenitor phenotype on colorectal cancer cells. Cell 111, 241–250 (2002).
Schwitalla, S. et al. Intestinal tumorigenesis initiated by dedifferentiation and acquisition of stem-cell-like properties. Cell 152, 25–38 (2013).
Brabletz, T. et al. Variable beta-catenin expression in colorectal cancers indicates tumor progression driven by the tumor environment. Proc. Natl. Acad. Sci. USA 98, 10356–10361 (2001).
Vermeulen, L. et al. Wnt activity defines colon cancer stem cells and is regulated by the microenvironment. Nat. Cell Biol. 12, 468–476 (2010).
de Sousa e Melo, F. et al. A distinct role for Lgr5+ stem cells in primary and metastatic colon cancer. Nature 543, 676–680 (2017).
Suvà, M.L. et al. Reconstructing and reprogramming the tumor-propagating potential of glioblastoma stem-like cells. Cell 157, 580–594 (2014).
Batlle, E. et al. The transcription factor snail is a repressor of E-cadherin gene expression in epithelial tumour cells. Nat. Cell Biol. 2, 84–89 (2000).
Cano, A. et al. The transcription factor snail controls epithelial-mesenchymal transitions by repressing E-cadherin expression. Nat. Cell Biol. 2, 76–83 (2000).
Savagner, P., Yamada, K.M. & Thiery, J.P. The zinc-finger protein slug causes desmosome dissociation, an initial and necessary step for growth factor-induced epithelial-mesenchymal transition. J. Cell Biol. 137, 1403–1419 (1997).
Mani, S.A. et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell 133, 704–715 (2008).
Morel, A.P. et al. Generation of breast cancer stem cells through epithelial-mesenchymal transition. PLoS One 3, e2888 (2008).
Puisieux, A., Brabletz, T. & Caramel, J. Oncogenic roles of EMT-inducing transcription factors. Nat. Cell Biol. 16, 488–494 (2014).
Ye, X. et al. Distinct EMT programs control normal mammary stem cells and tumour-initiating cells. Nature 525, 256–260 (2015).
Guo, W. et al. Slug and Sox9 cooperatively determine the mammary stem cell state. Cell 148, 1015–1028 (2012).
Nieto, M.A., Huang, R.Y., Jackson, R.A. & Thiery, J.P. EMT: 2016. Cell 166, 21–45 (2016).
Celià-Terrassa, T. et al. Epithelial-mesenchymal transition can suppress major attributes of human epithelial tumor-initiating cells. J. Clin. Invest. 122, 1849–1868 (2012).
Tran, H.D. et al. Transient SNAIL1 expression is necessary for metastatic competence in breast cancer. Cancer Res. 74, 6330–6340 (2014).
Tsai, J.H., Donaher, J.L., Murphy, D.A., Chau, S. & Yang, J. Spatiotemporal regulation of epithelial-mesenchymal transition is essential for squamous cell carcinoma metastasis. Cancer Cell 22, 725–736 (2012).
Ocaña, O.H. et al. Metastatic colonization requires the repression of the epithelial-mesenchymal transition inducer Prrx1. Cancer Cell 22, 709–724 (2012).
Beerling, E. et al. Plasticity between epithelial and mesenchymal states unlinks EMT from metastasis-enhancing stem cell capacity. Cell Reports 14, 2281–2288 (2016).
Beck, B. et al. Different levels of Twist1 regulate skin tumor initiation, stemness, and progression. Cell Stem Cell 16, 67–79 (2015).
Chaffer, C.L. et al. Poised chromatin at the ZEB1 promoter enables breast cancer cell plasticity and enhances tumorigenicity. Cell 154, 61–74 (2013).
Schmidt, J.M. et al. Stem-cell-like properties and epithelial plasticity arise as stable traits after transient Twist1 activation. Cell Reports 10, 131–139 (2015).
Suda, T., Takubo, K. & Semenza, G.L. Metabolic regulation of hematopoietic stem cells in the hypoxic niche. Cell Stem Cell 9, 298–310 (2011).
Simsek, T. et al. The distinct metabolic profile of hematopoietic stem cells reflects their location in a hypoxic niche. Cell Stem Cell 7, 380–390 (2010).
Maryanovich, M. et al. An MTCH2 pathway repressing mitochondria metabolism regulates haematopoietic stem cell fate. Nat. Commun. 6, 7901 (2015).
Tothova, Z. et al. FoxOs are critical mediators of hematopoietic stem cell resistance to physiologic oxidative stress. Cell 128, 325–339 (2007).
Ryall, J.G. et al. The NAD+-dependent SIRT1 deacetylase translates a metabolic switch into regulatory epigenetics in skeletal muscle stem cells. Cell Stem Cell 16, 171–183 (2015).
Rodríguez-Colman, M.J. et al. Interplay between metabolic identities in the intestinal crypt supports stem cell function. Nature 543, 424–427 (2017).
Yilmaz, O.H. et al. mTORC1 in the Paneth cell niche couples intestinal stem-cell function to calorie intake. Nature 486, 490–495 (2012).
Viale, A. et al. Oncogene ablation-resistant pancreatic cancer cells depend on mitochondrial function. Nature 514, 628–632 (2014).
Sancho, P. et al. MYC/PGC-1a balance determines the metabolic phenotype and plasticity of pancreatic cancer stem cells. Cell Metab. 22, 590–605 (2015).
Peiris-Pagès, M., Martinez-Outschoorn, U.E., Pestell, R.G., Sotgia, F. & Lisanti, M.P. Cancer stem cell metabolism. Breast Cancer Res. 18, 55 (2016).
Sancho, P., Barneda, D. & Heeschen, C. Hallmarks of cancer stem cell metabolism. Br. J. Cancer 114, 1305–1312 (2016).
Vlashi, E. et al. Metabolic state of glioma stem cells and nontumorigenic cells. Proc. Natl. Acad. Sci. USA 108, 16062–16067 (2011).
Flavahan, W.A. et al. Brain tumor initiating cells adapt to restricted nutrition through preferential glucose uptake. Nat. Neurosci. 16, 1373–1382 (2013).
Dong, C. et al. Loss of FBP1 by Snail-mediated repression provides metabolic advantages in basal-like breast cancer. Cancer Cell 23, 316–331 (2013).
Krebs, A.M. et al. The EMT-activator Zeb1 is a key factor for cell plasticity and promotes metastasis in pancreatic cancer. Nat. Cell Biol. 19, 518–529 (2017).
Dupuy, F. et al. PDK1-dependent metabolic reprogramming dictates metastatic potential in breast cancer. Cell Metab. 22, 577–589 (2015).
Pascual, G. et al. Targeting metastasis-initiating cells through the fatty acid receptor CD36. Nature 541, 41–45 (2017).
Ye, H. et al. Leukemic stem cells evade chemotherapy by metabolic adaptation to an adipose tissue niche. Cell Stem Cell 19, 23–37 (2016).
Borst, P. Cancer drug pan-resistance: pumps, cancer stem cells, quiescence, epithelial to mesenchymal transition, blocked cell death pathways, persisters or what? Open Biol. 2, 120066 (2012).
Holohan, C., Van Schaeybroeck, S., Longley, D.B. & Johnston, P.G. Cancer drug resistance: an evolving paradigm. Nat. Rev. Cancer 13, 714–726 (2013).
Diehn, M. et al. Association of reactive oxygen species levels and radioresistance in cancer stem cells. Nature 458, 780–783 (2009).
Li, X. et al. Intrinsic resistance of tumorigenic breast cancer cells to chemotherapy. J. Natl. Cancer Inst. 100, 672–679 (2008).
Bao, S. et al. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature 444, 756–760 (2006).
Clarkson, B., et al. Studies of cellular proliferation in human leukemia .3. Behavior of leukemic cells in three adults with acute leukemia given continuous infusions of 3H-thymidine for 8 or 10 days. Cancer 25, 1237–1260 (1970).
Killmann, S.A., Cronkite, E.P., Robertson, J.S., Fliedner, T.M. & Bond, V.P. Estimation of phases of the life cycle of leukemic cells from labeling in human beings in vivo with tritiated thymidine. Lab. Invest. 12, 671–684 (1963).
Clarkson, B.D. Review of recent studies of cellular proliferation in acute leukemia. Natl. Cancer Inst. Monogr. 30, 81–120 (1969).
Clarkson, B. The survival value of the dormant state in neoplastic and normal populations. in Control of Proliferation in Animal Cells. (eds. Clarkson, B., Baserga, R.) 945–972 (Cold Spring Harbor Laboratory, New York, NY, 1974).
Clarkson, B.D. & Fried, J. Changing concepts of treatment in acute leukemia. Med. Clin. North Am. 55, 561–600 (1971).
Cronkite, E.P. Acute leukemia: is there a relationship between cell growth kinetics and response to chemotherapy? Proc. Natl. Cancer Conf. 6, 113–117 (1970).
Kreso, A. et al. Variable clonal repopulation dynamics influence chemotherapy response in colorectal cancer. Science 339, 543–548 (2013).
Massagué, J. TGFb signalling in context. Nat. Rev. Mol. Cell Biol. 13, 616–630 (2012).
Bragado, P. et al. TGF-b2 dictates disseminated tumour cell fate in target organs through TGF-b-RIII and p38a/b signalling. Nat. Cell Biol. 15, 1351–1361 (2013).
Liau, B.B. et al. Adaptive chromatin remodeling drives glioblastoma stem cell plasticity and drug tolerance. Cell Stem Cell 20, 233–246 (2017).
Kurtova, A.V. et al. Blocking PGE2-induced tumour repopulation abrogates bladder cancer chemoresistance. Nature 517, 209–213 (2015).
Creighton, C.J. et al. Residual breast cancers after conventional therapy display mesenchymal as well as tumor-initiating features. Proc. Natl. Acad. Sci. USA 106, 13820–13825 (2009).
Roesch, A. et al. A temporarily distinct subpopulation of slow-cycling melanoma cells is required for continuous tumor growth. Cell 141, 583–594 (2010).
Roesch, A. et al. Overcoming intrinsic multidrug resistance in melanoma by blocking the mitochondrial respiratory chain of slow-cycling JARID1B(high) cells. Cancer Cell 23, 811–825 (2013).
Stange, D.E. et al. Differentiated Troy+ chief cells act as reserve stem cells to generate all lineages of the stomach epithelium. Cell 155, 357–368 (2013).
Wei, L. et al. Inhibition of CDK4/6 protects against radiation-induced intestinal injury in mice. J. Clin. Invest. 126, 4076–4087 (2016).
Nowak, D., Stewart, D. & Koeffler, H.P. Differentiation therapy of leukemia: 3 decades of development. Blood 113, 3655–3665 (2009).
Stahl, M. et al. Epigenetics in cancer: a hematological perspective. PLoS Genet. 12, e1006193 (2016).
Harris, W.J. et al. The histone demethylase KDM1A sustains the oncogenic potential of MLL-AF9 leukemia stem cells. Cancer Cell 21, 473–487 (2012).
Kreso, A. et al. Self-renewal as a therapeutic target in human colorectal cancer. Nat. Med. 20, 29–36 (2014).
Boumahdi, S. et al. SOX2 controls tumour initiation and cancer stem-cell functions in squamous-cell carcinoma. Nature 511, 246–250 (2014).
Junttila, M.R. et al. Targeting LGR5+ cells with an antibody-drug conjugate for the treatment of colon cancer. Sci. Transl. Med. 7, 314ra186 (2015).
Gong, X. et al. LGR5-targeted antibody-drug conjugate eradicates gastrointestinal tumors and prevents recurrence. Mol. Cancer Ther. 15, 1580–1590 (2016).
Saunders, L.R. et al. A DLL3-targeted antibody-drug conjugate eradicates high-grade pulmonary neuroendocrine tumor-initiating cells in vivo. Sci. Transl. Med. 7, 302ra136 (2015).
Takeishi, S. et al. Ablation of Fbxw7 eliminates leukemia-initiating cells by preventing quiescence. Cancer Cell 23, 347–361 (2013).
Reavie, L. et al. Regulation of c-Myc ubiquitination controls chronic myelogenous leukemia initiation and progression. Cancer Cell 23, 362–375 (2013).
Reya, T. & Clevers, H. Wnt signalling in stem cells and cancer. Nature 434, 843–850 (2005).
Anastas, J.N. & Moon, R.T. WNT signalling pathways as therapeutic targets in cancer. Nat. Rev. Cancer 13, 11–26 (2013).
Nile, A.H. & Hannoush, R.N. Fatty acylation of Wnt proteins. Nat. Chem. Biol. 12, 60–69 (2016).
Jiang, X. et al. Inactivating mutations of RNF43 confer Wnt dependency in pancreatic ductal adenocarcinoma. Proc. Natl. Acad. Sci. USA 110, 12649–12654 (2013).
Koo, B.K., van Es, J.H., van den Born, M. & Clevers, H. Porcupine inhibitor suppresses paracrine Wnt-driven growth of Rnf43;Znrf3-mutant neoplasia. Proc. Natl. Acad. Sci. USA 112, 7548–7550 (2015).
van de Wetering, M. et al. Prospective derivation of a living organoid biobank of colorectal cancer patients. Cell 161, 933–945 (2015).
Tammela, T. et al. A Wnt-producing niche drives proliferative potential and progression in lung adenocarcinoma. Nature 545, 355–359 (2017).
Steinhart, Z. et al. Genome-wide CRISPR screens reveal a Wnt-FZD5 signaling circuit as a druggable vulnerability of RNF43-mutant pancreatic tumors. Nat. Med. 23, 60–68 (2017).
Seshagiri, S. et al. Recurrent R-spondin fusions in colon cancer. Nature 488, 660–664 (2012).
Storm, E.E. et al. Targeting PTPRK-RSPO3 colon tumours promotes differentiation and loss of stem-cell function. Nature 529, 97–100 (2016).
Emami, K.H. et al. A small molecule inhibitor of beta-catenin/CREB-binding protein transcription [corrected]. Proc. Natl. Acad. Sci. USA 101, 12682–12687 (2004).
Novellasdemunt, L., Antas, P. & Li, V.S. Targeting Wnt signaling in colorectal cancer. A review in the theme: cell signaling: proteins, pathways and mechanisms. Am. J. Physiol. Cell Physiol. 309, C511–C521 (2015).
Kahn, M. Can we safely target the WNT pathway? Nat. Rev. Drug Discov. 13, 513–532 (2014).
Matano, M. et al. Modeling colorectal cancer using CRISPR-Cas9-mediated engineering of human intestinal organoids. Nat. Med. 21, 256–262 (2015).
Drost, J. et al. Sequential cancer mutations in cultured human intestinal stem cells. Nature 521, 43–47 (2015).
Fumagalli, A. et al. Genetic dissection of colorectal cancer progression by orthotopic transplantation of engineered cancer organoids. Proc. Natl. Acad. Sci. USA 114, E2357–E2364 (2017).
Fujii, M. et al. A colorectal tumor organoid library demonstrates progressive loss of niche factor requirements during tumorigenesis. Cell Stem Cell 18, 827–838 (2016).
Oskarsson, T. et al. Breast cancer cells produce tenascin C as a metastatic niche component to colonize the lungs. Nat. Med. 17, 867–874 (2011).
Malanchi, I. et al. Interactions between cancer stem cells and their niche govern metastatic colonization. Nature 481, 85–89 (2011).
McGranahan, N. & Swanton, C. Clonal heterogeneity and tumor evolution: past, present, and the future. Cell 168, 613–628 (2017).
Takebe, N. et al. Targeting Notch, Hedgehog, and Wnt pathways in cancer stem cells: clinical update. Nat. Rev. Clin. Oncol. 12, 445–464 (2015).
Andersson, E.R. & Lendahl, U. Therapeutic modulation of Notch signalling--are we there yet? Nat. Rev. Drug Discov. 13, 357–378 (2014).
Laszlo, G.S., Estey, E.H. & Walter, R.B. The past and future of CD33 as therapeutic target in acute myeloid leukemia. Blood Rev. 28, 143–153 (2014).
de Goeij, B.E. & Lambert, J.M. New developments for antibody-drug conjugate-based therapeutic approaches. Curr. Opin. Immunol. 40, 14–23 (2016).
Nervi, C., De Marinis, E. & Codacci-Pisanelli, G. Epigenetic treatment of solid tumours: a review of clinical trials. Clin. Epigenetics 7, 127 (2015).
Soriano, P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat. Genet. 21, 70–71 (1999).
Blanpain, C. & Simons, B.D. Unravelling stem cell dynamics by lineage tracing. Nat. Rev. Mol. Cell Biol. 14, 489–502 (2013).
Tata, P.R. & Rajagopal, J. Cellular plasticity: 1712 to the present day. Curr. Opin. Cell Biol. 43, 46–54 (2016).
Llorens-Bobadilla, E. & Martin-Villalba, A. Adult NSC diversity and plasticity: the role of the niche. Curr. Opin. Neurobiol. 42, 68–74 (2017).
Blanpain, C., Lowry, W.E., Geoghegan, A., Polak, L. & Fuchs, E. Self-renewal, multipotency, and the existence of two cell populations within an epithelial stem cell niche. Cell 118, 635–648 (2004).
Morris, R.J. et al. Capturing and profiling adult hair follicle stem cells. Nat. Biotechnol. 22, 411–417 (2004).
Ito, M. et al. Stem cells in the hair follicle bulge contribute to wound repair but not to homeostasis of the epidermis. Nat. Med. 11, 1351–1354 (2005).
Stingl, J. et al. Purification and unique properties of mammary epithelial stem cells. Nature 439, 993–997 (2006).
Shackleton, M. et al. Generation of a functional mammary gland from a single stem cell. Nature 439, 84–88 (2006).
Van Keymeulen, A. et al. Distinct stem cells contribute to mammary gland development and maintenance. Nature 479, 189–193 (2011).
Prater, M.D. et al. Mammary stem cells have myoepithelial cell properties. Nat. Cell Biol. 16, 942–950 (2014).
Rios, A.C., Fu, N.Y., Lindeman, G.J. & Visvader, J.E. In situ identification of bipotent stem cells in the mammary gland. Nature 506, 322–327 (2014).
Wuidart, A. et al. Quantitative lineage-tracing strategies to resolve multipotency in tissue-specific stem cells. Genes Dev. 30, 1261–1277 (2016).
Sun, J. et al. Clonal dynamics of native haematopoiesis. Nature 514, 322–327 (2014).
Busch, K. et al. Fundamental properties of unperturbed haematopoiesis from stem cells in vivo. Nature 518, 542–546 (2015).
Acknowledgements
Work in the laboratory of E.B. is supported by Fundación Botín and Banco Santander, through Santander Universities and the European Research Council (advanced grant no. 340176). We thank E. Sancho and J. Stanisavljevic for proofreading the manuscript.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Batlle, E., Clevers, H. Cancer stem cells revisited. Nat Med 23, 1124–1134 (2017). https://doi.org/10.1038/nm.4409
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nm.4409
This article is cited by
-
Cancer metabolism and carcinogenesis
Experimental Hematology & Oncology (2024)
-
Nanobiotechnology augmented cancer stem cell guided management of cancer: liquid-biopsy, imaging, and treatment
Journal of Nanobiotechnology (2024)
-
Essential role of PLD2 in hypoxia-induced stemness and therapy resistance in ovarian tumors
Journal of Experimental & Clinical Cancer Research (2024)
-
RACGAP1 promotes the progression and poor prognosis of lung adenocarcinoma through its effects on the cell cycle and tumor stemness
BMC Cancer (2024)
-
Crosstalk between colorectal CSCs and immune cells in tumorigenesis, and strategies for targeting colorectal CSCs
Experimental Hematology & Oncology (2024)