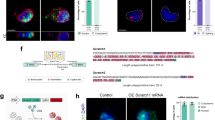
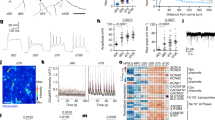
Abstract
Embryonal tumors with multilayered rosettes (ETMRs) have recently been described as a new entity of rare pediatric brain tumors with a fatal outcome. We show here that ETMRs are characterized by a parallel activation of Shh and Wnt signaling. Co-activation of these pathways in mouse neural precursors is sufficient to induce ETMR-like tumors in vivo that resemble their human counterparts on the basis of histology and global gene-expression analyses, and that point to apical radial glia cells as the possible tumor cell of origin. Overexpression of LIN28A, which is a hallmark of human ETMRs, augments Sonic-hedgehog (Shh) and Wnt signaling in these precursor cells through the downregulation of let7-miRNA, and LIN28A/let7a interaction with the Shh pathway was detected at the level of Gli mRNA. Finally, human ETMR cells that were transplanted into immunocompromised host mice were responsive to the SHH inhibitor arsenic trioxide (ATO). Our work provides a novel mouse model in which to study this tumor type, demonstrates the driving role of Wnt and Shh activation in the growth of ETMRs and proposes downstream inhibition of Shh signaling as a therapeutic option for patients with ETMRs.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Li, M. et al. Frequent amplification of a chr19q13.41 microRNA polycistron in aggressive primitive neuroectodermal brain tumors. Cancer Cell 16, 533–546 (2009).
Spence, T. et al. CNS-PNETs with C19MC amplification and/or LIN28 expression comprise a distinct histogenetic diagnostic and therapeutic entity. Acta Neuropathol. 128, 291–303 (2014).
Korshunov, A. et al. Focal genomic amplification at 19q13.42 comprises a powerful diagnostic marker for embryonal tumors with ependymoblastic rosettes. Acta Neuropathol. 120, 253–260 (2010).
Korshunov, A. et al. LIN28A immunoreactivity is a potent diagnostic marker of embryonal tumor with multilayered rosettes (ETMR). Acta Neuropathol. 124, 875–881 (2012).
Kleinman, C.L. et al. Fusion of TTYH1 with the C19MC microRNA cluster drives expression of a brain-specific DNMT3B isoform in the embryonal brain tumor ETMR. Nat. Genet. 46, 39–44 (2014).
Picard, D. et al. Markers of survival and metastatic potential in childhood CNS primitive neuro-ectodermal brain tumours: an integrative genomic analysis. Lancet Oncol. 13, 838–848 (2012).
Korshunov, A. et al. Embryonal tumor with abundant neuropil and true rosettes (ETANTR), ependymoblastoma, and medulloepithelioma share molecular similarity and comprise a single clinicopathological entity. Acta Neuropathol. 128, 279–289 (2014).
Sturm, D. et al. New brain tumor entities emerge from molecular classification of CNS-PNETs. Cell 164, 1060–1072 (2016).
Zurawel, R.H., Chiappa, S.A., Allen, C. & Raffel, C. Sporadic medulloblastomas contain oncogenic β-catenin mutations. Cancer Res. 58, 896–899 (1998).
Koch, A. et al. Somatic mutations of WNT/wingless signaling pathway components in primitive neuroectodermal tumors. Int. J. Cancer 93, 445–449 (2001).
Ishizaki, Y. et al. Immunohistochemical analysis and mutational analyses of beta-catenin, Axin family and APC genes in hepatocellular carcinomas. Int. J. Oncol. 24, 1077–1083 (2004).
Northcott, P.A. et al. Subgroup-specific structural variation across 1,000 medulloblastoma genomes. Nature 488, 49–56 (2012).
Al-Fageeh, M., Li, Q., Dashwood, W.M., Myzak, M.C. & Dashwood, R.H. Phosphorylation and ubiquitination of oncogenic mutants of beta-catenin containing substitutions at Asp32. Oncogene 23, 4839–4846 (2004).
Schwalbe, E.C. et al. Rapid diagnosis of medulloblastoma molecular subgroups. Clin. Cancer Res. 17, 1883–1894 (2011).
Kool, M. et al. Genome sequencing of SHH medulloblastoma predicts genotype-related response to smoothened inhibition. Cancer Cell 25, 393–405 (2014).
Zhuo, L. et al. hGFAP-cre transgenic mice for manipulation of glial and neuronal function in vivo. Genesis 31, 85–94 (2001).
Pöschl, J., Grammel, D., Dorostkar, M.M., Kretzschmar, H.A. & Schüller, U. Constitutive activation of β-catenin in neural progenitors results in disrupted proliferation and migration of neurons within the central nervous system. Dev. Biol. 374, 319–332 (2013).
Schüller, U. et al. Acquisition of granule neuron precursor identity is a critical determinant of progenitor cell competence to form Shh-induced medulloblastoma. Cancer Cell 14, 123–134 (2008).
Grammel, D. et al. Sonic hedgehog-associated medulloblastoma arising from the cochlear nuclei of the brainstem. Acta Neuropathol. 123, 601–614 (2012).
Pöschl, J. et al. Wnt/β-catenin signaling inhibits the Shh pathway and impairs tumor growth in Shh-dependent medulloblastoma. Acta Neuropathol. 127, 605–607 (2014).
Florio, M. & Huttner, W.B. Neural progenitors, neurogenesis and the evolution of the neocortex. Development 141, 2182–2194 (2014).
Florio, M. et al. Human-specific gene ARHGAP11B promotes basal progenitor amplification and neocortex expansion. Science 347, 1465–1470 (2015).
Spence, T. et al. A novel C19MC amplified cell line links Lin28/let-7 to mTOR signaling in embryonal tumor with multilayered rosettes. Neuro-oncol. 16, 62–71 (2014).
Pasca di Magliano, M. & Hebrok, M. Hedgehog signalling in cancer formation and maintenance. Nat. Rev. Cancer 3, 903–911 (2003).
Viswanathan, S.R., Daley, G.Q. & Gregory, R.I. Selective blockade of microRNA processing by Lin28. Science 320, 97–100 (2008).
Bagga, S. et al. Regulation by let-7 and lin-4 miRNAs results in target mRNA degradation. Cell 122, 553–563 (2005).
Emami, K.H. et al. A small molecule inhibitor of β-catenin/CREB-binding protein transcription [corrected]. Proc. Natl. Acad. Sci. USA 101, 12682–12687 (2004).
Dierks, C. GDC-0449—targeting the hedgehog signaling pathway. Recent Results Cancer Res. 184, 235–238 (2010).
Beauchamp, E.M. et al. Arsenic trioxide inhibits human cancer cell growth and tumor development in mice by blocking Hedgehog/GLI pathway. J. Clin. Invest. 121, 148–160 (2011).
Kim, J., Lee, J.J., Kim, J., Gardner, D. & Beachy, P.A. Arsenic antagonizes the Hedgehog pathway by preventing ciliary accumulation and reducing stability of the Gli2 transcriptional effector. Proc. Natl. Acad. Sci. USA 107, 13432–13437 (2010).
Chenn, A. & Walsh, C.A. Regulation of cerebral cortical size by control of cell cycle exit in neural precursors. Science 297, 365–369 (2002).
Nowak, J. et al. Systematic comparison of MRI findings in pediatric ependymoblastoma with ependymoma and CNS primitive neuroectodermal tumor not otherwise specified. Neuro-oncol. 17, 1157–1165 (2015).
Noctor, S.C., Martínez-Cerdeño, V., Ivic, L. & Kriegstein, A.R. Cortical neurons arise in symmetric and asymmetric division zones and migrate through specific phases. Nat. Neurosci. 7, 136–144 (2004).
Sato, H. et al. Embryonal tumor with abundant neuropil and true rosettes in the brainstem: case report. J. Neurosurg. Pediatr. 16, 291–295 (2015).
Nowak, J. et al. MRI characteristics of ependymoblastoma: results from 22 centrally reviewed cases. AJNR Am. J. Neuroradiol. 35, 1996–2001 (2014).
Gessi, M. et al. Embryonal tumors with abundant neuropil and true rosettes: a distinctive CNS primitive neuroectodermal tumor. Am. J. Surg. Pathol. 33, 211–217 (2009).
Yang, M. et al. Lin28 promotes the proliferative capacity of neural progenitor cells in brain development. Development 142, 1616–1627 (2015).
Bentwich, I. et al. Identification of hundreds of conserved and nonconserved human microRNAs. Nat. Genet. 37, 766–770 (2005).
Lei, X.X. et al. Determinants of mRNA recognition and translation regulation by Lin28. Nucleic Acids Res. 40, 3574–3584 (2012).
Weingart, M.F. et al. Disrupting LIN28 in atypical teratoid rhabdoid tumors reveals the importance of the mitogen activated protein kinase pathway as a therapeutic target. Oncotarget 6, 3165–3177 (2015).
Zeng, Y. et al. Lin28A binds active promoters and recruits Tet1 to regulate gene expression. Mol. Cell 61, 153–160 (2016).
Schmidt, C. et al. Pre-clinical drug screen reveals topotecan, actinomycin D and volasertib as potential new therapeutic candidates for ETMR brain tumor patients. Neuro Oncol. http://dx.doi.org/10.1093/neuonc/nox093 (2017).
Iland, H.J. et al. Use of arsenic trioxide in remission induction and consolidation therapy for acute promyelocytic leukaemia in the Australasian Leukaemia and Lymphoma Group (ALLG) APML4 study: a non-randomised phase 2 trial. Lancet Haematol. 2, e357–e366 (2015).
Sturm, D. et al. Hotspot mutations in H3F3A and IDH1 define distinct epigenetic and biological subgroups of glioblastoma. Cancer Cell 22, 425–437 (2012).
Pajtler, K.W. et al. Molecular classification of ependymal tumors across all CNS compartments, histopathological grades, and age groups. Cancer Cell 27, 728–743 (2015).
Arnold, K. et al. Sox2+ adult stem and progenitor cells are important for tissue regeneration and survival of mice. Cell Stem Cell 9, 317–329 (2011).
Mao, J. et al. A novel somatic mouse model to survey tumorigenic potential applied to the Hedgehog pathway. Cancer Res. 66, 10171–10178 (2006).
Shultz, L.D. et al. Human lymphoid and myeloid cell development in NOD/LtSz-scid IL2R gamma null mice engrafted with mobilized human hemopoietic stem cells. J. Immunol. 174, 6477–6489 (2005).
Harada, N. et al. Intestinal polyposis in mice with a dominant stable mutation of the beta-catenin gene. EMBO J. 18, 5931–5942 (1999).
Papaioannou, G., Inloes, J.B., Nakamura, Y., Paltrinieri, E. & Kobayashi, T. let-7 and miR-140 microRNAs coordinately regulate skeletal development. Proc. Natl. Acad. Sci. USA 110, E3291–E3300 (2013).
Hovestadt, V. et al. Robust molecular subgrouping and copy-number profiling of medulloblastoma from small amounts of archival tumour material using high-density DNA methylation arrays. Acta Neuropathol. 125, 913–916 (2013).
Nowak, J. et al. Ependymoblastoma of the brainstem: MRI findings and differential diagnosis. Pediatr. Blood Cancer 61, 1132–1134 (2014).
Friedrich, C. et al. Primitive neuroectodermal tumors of the brainstem in children treated according to the HIT trials: clinical findings of a rare disease. J. Neurosurg. Pediatr. 15, 227–235 (2015).
Stock, K. et al. Neural precursor cells induce cell death of high-grade astrocytomas through stimulation of TRPV1. Nat. Med. 18, 1232–1238 (2012).
Svärd, J. et al. Genetic elimination of Suppressor of fused reveals an essential repressor function in the mammalian Hedgehog signaling pathway. Dev. Cell 10, 187–197 (2006).
Sinha, S. & Chen, J.K. Purmorphamine activates the Hedgehog pathway by targeting Smoothened. Nat. Chem. Biol. 2, 29–30 (2006).
Lipinski, R.J., Bijlsma, M.F., Gipp, J.J., Podhaizer, D.J. & Bushman, W. Establishment and characterization of immortalized Gli-null mouse embryonic fibroblast cell lines. BMC Cell Biol. 9, 49 (2008).
Kingston, R.E., Chen, C.A. & Okayama, H. Calcium phosphate transfection. Curr. Protoc. Cell Biol. Chapter 20, Unit 20.23 (2003).
Wani, S. & Cloonan, N. Profiling direct mRNA-microRNA interactions using synthetic biotinylated microRNA-duplexes. Preprint at https://doi.org/10.1101/005439 (2014).
Schüller, U. et al. Forkhead transcription factor FoxM1 regulates mitotic entry and prevents spindle defects in cerebellar granule neuron precursors. Mol. Cell. Biol. 27, 8259–8270 (2007).
Pöschl, J. et al. Expression of BARHL1 in medulloblastoma is associated with prolonged survival in mice and humans. Oncogene 30, 4721–4730 (2011).
Hovestadt, V. et al. Decoding the regulatory landscape of medulloblastoma using DNA methylation sequencing. Nature 510, 537–541 (2014).
Jones, D.T. et al. Dissecting the genomic complexity underlying medulloblastoma. Nature 488, 100–105 (2012).
Sahm, F. et al. Next-generation sequencing in routine brain tumor diagnostics enables an integrated diagnosis and identifies actionable targets. Acta Neuropathol. 131, 903–910 (2016).
Yang, H. & Wang, K. Genomic variant annotation and prioritization with ANNOVAR and wANNOVAR. Nat. Protoc. 10, 1556–1566 (2015).
Robinson, G. et al. Novel mutations target distinct subgroups of medulloblastoma. Nature 488, 43–48 (2012).
Lambert, N. et al. Genes expressed in specific areas of the human fetal cerebral cortex display distinct patterns of evolution. PLoS One 6, e17753 (2011).
Pöschl, J. et al. Genomic and transcriptomic analyses match medulloblastoma mouse models to their human counterparts. Acta Neuropathol. 128, 123–136 (2014).
Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 102, 15545–15550 (2005).
Kanehisa, M. & Goto, S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28, 27–30 (2000).
Shannon, P. et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504 (2003).
Acknowledgements
We thank T. Kobayashi (Harvard University, Boston, USA) for providing the LIN28A(3x)-IRES-eGFP plasmid, D. Rowitch (University of Cambridge, Cambridge, USA) for providing MSCV-IRES-GFP and MSCV-Cre-IRES-GFP plasmids, and D. Baltimore (Pasadena, CA, USA) for providing the Lin28A-IRES-GFP plasmid. We thank R. Toftgård, (Karolinska Institute, Stockholm, Sweden), for providing Sufu−/− MEFs, J. Chen (Stanford University School of Medicine, Stanford, USA) for providing Smo−/− MEFs, and R. Lipinski (University of Wisconsin, Madison, USA) for providing Gli1−/−, Gli2−/−, and Gli3−/− MEFs. We thank H. Blum and S. Krebs for assistance with murine gene-expression data (Gene Center Munich, Germany). We thank S. Occhionero, M. Burmester, M. Wagner, and M. Schmidt (LMU, Munich, Germany) and M. Gregersen and I. Nachtigall (UKE Hamburg, Germany) for excellent technical support, and P. Bonert, P. Liebmann, C. Mann, and M. Wellisch (LMU, Munich, Germany) for animal husbandry. We thank K. Hartmann from the mouse pathology core facility (UKE Hamburg, Germany) for processing immunohistochemical stainings. We are indebted to all members of the Schüller group for very fruitful discussions. This work was supported by the Fördergemeinschaft Kinderkrebs-Zentrum Hamburg and grants from the Deutsche Krebshilfe (Max-Eder junior research program to U.S.), the Wilhelm Sander Foundation (to U.S.), the Else-Kröner-Fresenius Foundation (to U.S. and J.E.N.), the K.L. Weigand foundation (to J.E.N.) and the Association “Förderung von Wissenschaft und Forschung an der Medizinischen Fakultät der LMU München e.V.” (to J.E.N.).
Author information
Authors and Affiliations
Contributions
J.E.N., A.K.W., and U.S. conceived the project and wrote the manuscript. J.E.N and A.K.W. conducted the majority of experiments. E.B., M.B., V.M., P.S., and M.S. performed experiments. J.N., P.N., L.C., T.S., and M.M.D. performed computational analysis on large-scale data; R.G., M.M.T., J.A.C., M.R.S., I.R-M., and D.J.M. provided assistance with mouse experiments and cell lines. S.L., A.K., and M.K. generated the sequencing data. M.K. generated human miRNA-seq data. K.v.H., J.N., and M.W.-M. provided assistance with human MRI data.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–12 and Supplementary Table 1 (PDF 15663 kb)
Supplementary Table 1
Supplementary Table 1 (XLSX 1967 kb)
Rights and permissions
About this article
Cite this article
Neumann, J., Wefers, A., Lambo, S. et al. A mouse model for embryonal tumors with multilayered rosettes uncovers the therapeutic potential of Sonic-hedgehog inhibitors. Nat Med 23, 1191–1202 (2017). https://doi.org/10.1038/nm.4402
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nm.4402
This article is cited by
-
A DEAD-box helicase drives the partitioning of a pro-differentiation NAB protein into nuclear foci
Nature Communications (2023)
-
Emerging role of non-coding RNAs in the regulation of Sonic Hedgehog signaling pathway
Cancer Cell International (2022)
-
Adult neurogenesis of the median eminence contributes to structural reconstruction and recovery of body fluid metabolism in hypothalamic self-repair after pituitary stalk lesion
Cellular and Molecular Life Sciences (2022)
-
Overexpression of Lin28A in neural progenitor cells in vivo does not lead to brain tumor formation but results in reduced spine density
Acta Neuropathologica Communications (2021)
-
Co-activation of Sonic hedgehog and Wnt signaling in murine retinal precursor cells drives ocular lesions with features of intraocular medulloepithelioma
Oncogenesis (2021)