Abstract
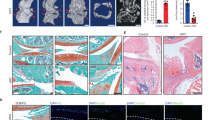
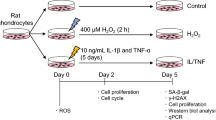
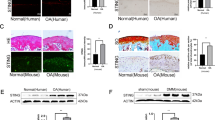
Senescent cells (SnCs) accumulate in many vertebrate tissues with age and contribute to age-related pathologies1,2,3, presumably through their secretion of factors contributing to the senescence-associated secretory phenotype (SASP)4,5,6. Removal of SnCs delays several pathologies7,8,9 and increases healthy lifespan8. Aging and trauma are risk factors for the development of osteoarthritis (OA)10, a chronic disease characterized by degeneration of articular cartilage leading to pain and physical disability. Senescent chondrocytes are found in cartilage tissue isolated from patients undergoing joint replacement surgery11,12,13,14, yet their role in disease pathogenesis is unknown. To test the idea that SnCs might play a causative role in OA, we used the p16-3MR transgenic mouse, which harbors a p16INK4a (Cdkn2a) promoter driving the expression of a fusion protein containing synthetic Renilla luciferase and monomeric red fluorescent protein domains, as well as a truncated form of herpes simplex virus 1 thymidine kinase (HSV-TK)15,16. This mouse strain allowed us to selectively follow and remove SnCs after anterior cruciate ligament transection (ACLT). We found that SnCs accumulated in the articular cartilage and synovium after ACLT, and selective elimination of these cells attenuated the development of post-traumatic OA, reduced pain and increased cartilage development. Intra-articular injection of a senolytic molecule that selectively killed SnCs validated these results in transgenic, non-transgenic and aged mice. Selective removal of the SnCs from in vitro cultures of chondrocytes isolated from patients with OA undergoing total knee replacement decreased expression of senescent and inflammatory markers while also increasing expression of cartilage tissue extracellular matrix proteins. Collectively, these findings support the use of SnCs as a therapeutic target for treating degenerative joint disease.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Campisi, J. Aging, cellular senescence, and cancer. Annu. Rev. Physiol. 75, 685–705 (2013).
van Deursen, J.M. The role of senescent cells in ageing. Nature 509, 439–446 (2014).
Campisi, J. Senescent cells, tumor suppression, and organismal aging: good citizens, bad neighbors. Cell 120, 513–522 (2005).
Coppé, J.P. et al. Senescence-associated secretory phenotypes reveal cell-nonautonomous functions of oncogenic RAS and the p53 tumor suppressor. PLoS Biol. 6, 2853–2868 (2008).
Campisi, J. Cancer, aging and cellular senescence. In Vivo 14, 183–188 (2000).
Nelson, G. et al. A senescent cell bystander effect: senescence-induced senescence. Aging Cell 11, 345–349 (2012).
Baker, D.J. et al. Clearance of p16Ink4a-positive senescent cells delays ageing-associated disorders. Nature 479, 232–236 (2011).
Baker, D.J. et al. Naturally occurring p16Ink4a-positive cells shorten healthy lifespan. Nature 530, 184–189 (2016).
Baker, D.J. et al. Opposing roles for p16Ink4a and p19Arf in senescence and ageing caused by BubR1 insufficiency. Nat. Cell Biol. 10, 825–836 (2008).
Wieland, H.A., Michaelis, M., Kirschbaum, B.J. & Rudolphi, K.A. Osteoarthritis—an untreatable disease? Nat. Rev. Drug Discov. 4, 331–344 (2005).
Martin, J.A., Brown, T., Heiner, A. & Buckwalter, J.A. Post-traumatic osteoarthritis: the role of accelerated chondrocyte senescence. Biorheology 41, 479–491 (2004).
Price, J.S. et al. The role of chondrocyte senescence in osteoarthritis. Aging Cell 1, 57–65 (2002).
Philipot, D. et al. p16INK4a and its regulator miR-24 link senescence and chondrocyte terminal differentiation–associated matrix remodeling in osteoarthritis. Arthritis Res. Ther. 16, R58 (2014).
McCulloch, K., Litherland, G.J. & Rai, T.S. Cellular senescence in osteoarthritis pathology. Aging Cell 16, 210–218 (2017).
Demaria, M. et al. An essential role for senescent cells in optimal wound healing through secretion of PDGF-AA. Dev. Cell 31, 722–733 (2014).
Chang, J. et al. Clearance of senescent cells by ABT263 rejuvenates aged hematopoietic stem cells in mice. Nat. Med. 22, 78–83 (2016).
Adams, P.D. Healing and hurting: molecular mechanisms, functions, and pathologies of cellular senescence. Mol. Cell 36, 2–14 (2009).
Sharpless, N.E. & Sherr, C.J. Forging a signature of in vivo senescence. Nat. Rev. Cancer 15, 397–408 (2015).
Ohtani, N., Yamakoshi, K., Takahashi, A. & Hara, E. The p16INK4a–RB pathway: molecular link between cellular senescence and tumor suppression. J. Med. Invest. 51, 146–153 (2004).
Salama, R., Sadaie, M., Hoare, M. & Narita, M. Cellular senescence and its effector programs. Genes Dev. 28, 99–114 (2014).
Holmlund, U. et al. The novel inflammatory cytokine high mobility group box protein 1 (HMGB1) is expressed by human term placenta. Immunology 122, 430–437 (2007).
Davalos, A.R. et al. p53-dependent release of alarmin HMGB1 is a central mediator of senescent phenotypes. J. Cell Biol. 201, 613–629 (2013).
Sellam, J. & Berenbaum, F. The role of synovitis in pathophysiology and clinical symptoms of osteoarthritis. Nat. Rev. Rheumatol. 6, 625–635 (2010).
Dowthwaite, G.P. et al. The surface of articular cartilage contains a progenitor cell population. J. Cell Sci. 117, 889–897 (2004).
Sharma, B. et al. Human cartilage repair with a photoreactive adhesive–hydrogel composite. Sci. Transl. Med. 5, 167ra6 (2013).
Laberge, R.M. et al. Mitochondrial DNA damage induces apoptosis in senescent cells. Cell Death Dis. 4, e727 (2013).
Goldring, M.B. & Otero, M. Inflammation in osteoarthritis. Curr. Opin. Rheumatol. 23, 471–478 (2011).
Burr, D.B. & Gallant, M.A. Bone remodelling in osteoarthritis. Nat. Rev. Rheumatol. 8, 665–673 (2012).
Zhu, Y. et al. Identification of a novel senolytic agent, navitoclax, targeting the Bcl-2 family of anti-apoptotic factors. Aging Cell 15, 428–435 (2016).
Komori, T. Signaling networks in RUNX2-dependent bone development. J. Cell. Biochem. 112, 750–755 (2011).
Ruan, M.Z. et al. Proteoglycan 4 expression protects against the development of osteoarthritis. Sci. Transl. Med. 5, 176ra34 (2013).
Loeser, R.F. et al. Microarray analysis reveals age-related differences in gene expression during the development of osteoarthritis in mice. Arthritis Rheum. 64, 705–717 (2012).
Loeser, R.F. Aging and osteoarthritis: the role of chondrocyte senescence and aging changes in the cartilage matrix. Osteoarthritis Cartilage 17, 971–979 (2009).
Sekiya, I., Vuoristo, J.T., Larson, B.L. & Prockop, D.J. In vitro cartilage formation by human adult stem cells from bone marrow stroma defines the sequence of cellular and molecular events during chondrogenesis. Proc. Natl. Acad. Sci. USA 99, 4397–4402 (2002).
Glasson, S.S., Chambers, M.G., Van Den Berg, W.B. & Little, C.B. The OARSI histopathology initiative—recommendations for histological assessments of osteoarthritis in the mouse. Osteoarthritis Cartilage 18 (Suppl. 3), S17–S23 (2010).
Schreiber, S., Backer, M.M., Yanai, J. & Pick, C.G. The antinociceptive effect of fluvoxamine. Eur. Neuropsychopharmacol. 6, 281–284 (1996).
Acknowledgements
We thank J. Xu (F.M. Kirby Research Center at Johns Hopkins University) for in vivo luminescence imaging, A. Bendele (Bolder Biopath, Inc.) for the subchondral bone damage analysis and Y. Oh (Johns Hopkins University) for immunoblotting. This work was supported by Unity Biotechnology, Inc. (J.H.E., A.P.V., Y.P., N.D.), the Bloomberg-Kimmel Institute for Cancer Immunotherapy (J.H.E.), the Morton Goldberg Professorship (J.H.E.), National Institute on Aging (NIA) grant AG017242 (J.C.), AG009909 (M.D.), National Cancer Institute (NCI) grant R01CA96985 (J.M.v.D.), a grant from the Paul F. Glenn Foundation (J.M.v.D. and D.J.B.) and a Fulbright scholarship from the Institute of International Education (O.H.J.).
Author information
Authors and Affiliations
Contributions
O.H.J. and C.K. designed, carried out and analyzed data from most of the experiments and wrote the manuscript with input from all co-authors; S.R. performed experiments; R.-M.L. and M.D. designed experiments and interpreted data; A.P.V. designed and analyzed data from experiments; J.C. provided mice, designed experiments, analyzed and interpreted data, and revised the manuscript; J.W.C. and D.H.K. performed experiments; Y.P. and N.D. conceived the application of senescence removal to OA treatment and participated in in vivo experimental design; D.J.B. and J.M.v.D. carried out experiments on naturally occurring OA; J.H.E. conceived, designed and supervised the study, analyzed and interpreted data, and wrote the manuscript. All authors discussed the results and commented on the manuscript.
Corresponding author
Ethics declarations
Competing interests
J.C., R.-M.L., Y.P., D.J.B. , J.M.v.D. , M.D., N.D. and J.H.E. own equity in Unity Biotechnology. Johns Hopkins University and Unity Biotechnology own intellectual property related to the research. O.H.J., C.K. and J.H.E. are inventors of Johns Hopkins University intellectual property licensed to Unity Biotechnology.
Supplementary information
Supplementary Figures and Table
Supplementary Figures 1–13 and Supplementary Table 1. (PDF 4267 kb)
Rights and permissions
About this article
Cite this article
Jeon, O., Kim, C., Laberge, RM. et al. Local clearance of senescent cells attenuates the development of post-traumatic osteoarthritis and creates a pro-regenerative environment. Nat Med 23, 775–781 (2017). https://doi.org/10.1038/nm.4324
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nm.4324
This article is cited by
-
Targeting an inflammation-amplifying cell population can attenuate osteoarthritis-associated pain
Arthritis Research & Therapy (2024)
-
Engineering approaches for RNA-based and cell-based osteoarthritis therapies
Nature Reviews Rheumatology (2024)
-
H3K36 methyltransferase NSD1 protects against osteoarthritis through regulating chondrocyte differentiation and cartilage homeostasis
Cell Death & Differentiation (2024)
-
Profiling joint tissues at single-cell resolution: advances and insights
Nature Reviews Rheumatology (2024)
-
Interferon-gamma signaling promotes cartilage regeneration after injury
Scientific Reports (2024)