Abstract
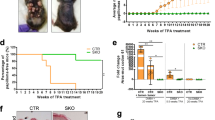
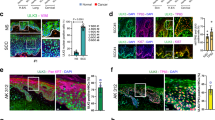
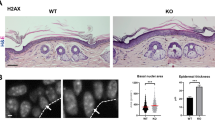
In a search for mediators of the p53 tumor suppressor pathway, which induces pleiotropic and often antagonistic cellular responses, we identified the long noncoding RNA (lncRNA) NEAT1. NEAT1 is an essential architectural component of paraspeckle nuclear bodies, whose pathophysiological relevance remains unclear. Activation of p53, pharmacologically or by oncogene-induced replication stress, stimulated the formation of paraspeckles in mouse and human cells. Silencing Neat1 expression in mice, which prevents paraspeckle formation, sensitized preneoplastic cells to DNA-damage-induced cell death and impaired skin tumorigenesis. We provide mechanistic evidence that NEAT1 promotes ATR signaling in response to replication stress and is thereby engaged in a negative feedback loop that attenuates oncogene-dependent activation of p53. NEAT1 targeting in established human cancer cell lines induced synthetic lethality with genotoxic chemotherapeutics, including PARP inhibitors, and nongenotoxic activation of p53. This study establishes a key genetic link between NEAT1 paraspeckles, p53 biology and tumorigenesis and identifies NEAT1 as a promising target to enhance sensitivity of cancer cells to both chemotherapy and p53 reactivation therapy.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Change history
08 February 2024
A Correction to this paper has been published: https://doi.org/10.1038/s41591-024-02842-w
References
Vogelstein, B., Lane, D. & Levine, A.J. Surfing the p53 network. Nature 408, 307–310 (2000).
Sharpless, N.E. & DePinho, R.A. How stem cells age and why this makes us grow old. Nat. Rev. Mol. Cell Biol. 8, 703–713 (2007).
Vassilev, L.T. et al. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science 303, 844–848 (2004).
Khoo, K.H., Verma, C.S. & Lane, D.P. Drugging the p53 pathway: understanding the route to clinical efficacy. Nat. Rev. Drug Discov. 13, 217–236 (2014).
Carvajal, L.A. & Manfredi, J.J. Another fork in the road–life or death decisions by the tumour suppressor p53. EMBO Rep. 14, 414–421 (2013).
Huang, B., Deo, D., Xia, M. & Vassilev, L.T. Pharmacologic p53 activation blocks cell cycle progression but fails to induce senescence in epithelial cancer cells. Mol. Cancer Res. 7, 1497–1509 (2009).
Derrien, T. et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 22, 1775–1789 (2012).
Yildirim, E. et al. Xist RNA is a potent suppressor of hematologic cancer in mice. Cell 152, 727–742 (2013).
Arun, G. et al. Differentiation of mammary tumors and reduction in metastasis upon Malat1 lncRNA loss. Genes Dev. 30, 34–51 (2016).
Huarte, M. et al. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell 142, 409–419 (2010).
Hung, T. et al. Extensive and coordinated transcription of noncoding RNAs within cell-cycle promoters. Nat. Genet. 43, 621–629 (2011).
Marín-Béjar, O. et al. Pint lincRNA connects the p53 pathway with epigenetic silencing by the Polycomb repressive complex 2. Genome Biol. 14, R104 (2013).
Blume, C.J. et al. p53-dependent non-coding RNA networks in chronic lymphocytic leukemia. Leukemia 29, 2015–2023 (2015).
Janky, R. et al. iRegulon: from a gene list to a gene regulatory network using large motif and track collections. PLoS Comput. Biol. 10, e1003731 (2014).
Herrmann, C., Van de Sande, B., Potier, D. & Aerts, S. i-cisTarget: an integrative genomics method for the prediction of regulatory features and cis-regulatory modules. Nucleic Acids Res. 40, e114 (2012).
Clemson, C.M. et al. An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol. Cell 33, 717–726 (2009).
Wilusz, J.E. et al. A triple helix stabilizes the 3′ ends of long noncoding RNAs that lack poly(A) tails. Genes Dev. 26, 2392–2407 (2012).
Chen, L.-L. & Carmichael, G.G. Altered nuclear retention of mRNAs containing inverted repeats in human embryonic stem cells: functional role of a nuclear noncoding RNA. Mol. Cell 35, 467–478 (2009).
Sasaki, Y.T.F., Ideue, T., Sano, M., Mituyama, T. & Hirose, T. MENepsilon/beta noncoding RNAs are essential for structural integrity of nuclear paraspeckles. Proc. Natl. Acad. Sci. USA 106, 2525–2530 (2009).
Sunwoo, H. et al. MEN-ɛ/β nuclear-retained non-coding RNAs are up-regulated upon muscle differentiation and are essential components of paraspeckles. Genome Res. 19, 347–359 (2009).
Yamazaki, T. & Hirose, T. The building process of the functional paraspeckle with long non-coding RNAs. Front. Biosci. (Elite Ed.) 7, 1–41 (2015).
Reinhardt, H.C. & Schumacher, B. The p53 network: cellular and systemic DNA damage responses in aging and cancer. Trends Genet. 28, 128–136 (2012).
Nassar, D., Latil, M., Boeckx, B., Lambrechts, D. & Blanpain, C. Genomic landscape of carcinogen-induced and genetically induced mouse skin squamous cell carcinoma. Nat. Med. 21, 946–954 (2015).
Lapouge, G. et al. Identifying the cellular origin of squamous skin tumors. Proc. Natl. Acad. Sci. USA 108, 7431–7436 (2011).
Nakagawa, S., Naganuma, T., Shioi, G. & Hirose, T. Paraspeckles are subpopulation-specific nuclear bodies that are not essential in mice. J. Cell Biol. 193, 31–39 (2011).
Gaillard, H., García-Muse, T. & Aguilera, A. Replication stress and cancer. Nat. Rev. Cancer 15, 276–289 (2015).
Llopis, A. et al. The stress-activated protein kinases p38α/β and JNK1/2 cooperate with Chk1 to inhibit mitotic entry upon DNA replication arrest. Cell Cycle 11, 3627–3637 (2012).
Branzei, D. & Foiani, M. Maintaining genome stability at the replication fork. Nat. Rev. Mol. Cell Biol. 11, 208–219 (2010).
Dobbelstein, M. & Sørensen, C.S. Exploiting replicative stress to treat cancer. Nat. Rev. Drug Discov. 14, 405–423 (2015).
Sullivan, K.D. et al. ATM and MET kinases are synthetic lethal with nongenotoxic activation of p53. Nat. Chem. Biol. 8, 646–654 (2012).
Ferriss, J.S. et al. Multi-gene expression predictors of single drug responses to adjuvant chemotherapy in ovarian carcinoma: predicting platinum resistance. PLoS One 7, e30550 (2012).
The Cancer Genome Atlas Research Network (TCGA). Integrated genomic analyses of ovarian carcinoma. Nature 474, 609–615 (2011).
Chakravarty, D. et al. The oestrogen receptor-α-regulated lncRNA NEAT1 is a critical modulator of prostate cancer. Nat. Commun. 5, 5383 (2014).
Choudhry, H. et al. Tumor hypoxia induces nuclear paraspeckle formation through HIF-2α dependent transcriptional activation of NEAT1 leading to cancer cell survival. Oncogene 34, 4482–4490 (2015).
Zhen, L. et al. Long noncoding RNA NEAT1 promotes glioma pathogenesis by regulating miR-449b-5p/c-Met axis. Tumour Biol. 37, 673–683 (2016).
Guo, S. et al. Clinical implication of long non-coding RNA NEAT1 expression in hepatocellular carcinoma patients. Int. J. Clin. Exp. Pathol. 8, 5395–5402 (2015).
Pan, L.-J. et al. Upregulation and clinicopathological significance of long non-coding NEAT1 RNA in NSCLC tissues. Asian Pac. J. Cancer Prev. 16, 2851–2855 (2015).
Sengupta, S. & Harris, C.C. p53: traffic cop at the crossroads of DNA repair and recombination. Nat. Rev. Mol. Cell Biol. 6, 44–55 (2005).
Harris, S.L. & Levine, A.J. The p53 pathway: positive and negative feedback loops. Oncogene 24, 2899–2908 (2005).
Cha, H. et al. Wip1 directly dephosphorylates γ-H2AX and attenuates the DNA damage response. Cancer Res. 70, 4112–4122 (2010).
Moon, S.-H. et al. Wild-type p53-induced phosphatase 1 dephosphorylates histone variant γ-H2AX and suppresses DNA double strand break repair. J. Biol. Chem. 285, 12935–12947 (2010).
Chowdhury, D. et al. γ-H2AX dephosphorylation by protein phosphatase 2A facilitates DNA double-strand break repair. Mol. Cell 20, 801–809 (2005).
Rulten, S.L. et al. PARP-1 dependent recruitment of the amyotrophic lateral sclerosis-associated protein FUS/TLS to sites of oxidative DNA damage. Nucleic Acids Res. 42, 307–314 (2014).
Krietsch, J. et al. PARP activation regulates the RNA-binding protein NONO in the DNA damage response to DNA double-strand breaks. Nucleic Acids Res. 40, 10287–10301 (2012).
Ha, K., Takeda, Y. & Dynan, W.S. Sequences in PSF/SFPQ mediate radioresistance and recruitment of PSF/SFPQ-containing complexes to DNA damage sites in human cells. DNA Repair (Amst.) 10, 252–259 (2011).
Salton, M., Lerenthal, Y., Wang, S.Y., Chen, D.J. & Shiloh, Y. Involvement of Matrin 3 and SFPQ/NONO in the DNA damage response. Cell Cycle 9, 1568–1576 (2010).
Mastrocola, A.S., Kim, S.H., Trinh, A.T., Rodenkirch, L.A. & Tibbetts, R.S. The RNA-binding protein fused in sarcoma (FUS) functions downstream of poly(ADP-ribose) polymerase (PARP) in response to DNA damage. J. Biol. Chem. 288, 24731–24741 (2013).
Altmeyer, M. et al. Liquid demixing of intrinsically disordered proteins is seeded by poly(ADP-ribose). Nat. Commun. 6, 8088 (2015).
Wickramasinghe, V.O. & Venkitaraman, A.R. RNA processing and genome stability: cause and consequence. Mol. Cell 61, 496–505 (2016).
Britton, S. et al. DNA damage triggers SAF-A and RNA biogenesis factors exclusion from chromatin coupled to R-loops removal. Nucleic Acids Res. 42, 9047–9062 (2014).
Shanbhag, N.M., Rafalska-Metcalf, I.U., Balane-Bolivar, C., Janicki, S.M. & Greenberg, R.A. ATM-dependent chromatin changes silence transcription in cis to DNA double-strand breaks. Cell 141, 970–981 (2010).
Specks, J., Lecona, E., Lopez-Contreras, A.J. & Fernandez-Capetillo, O. A single conserved residue mediates binding of the ribonucleotide reductase catalytic subunit RRM1 to RRM2 and is essential for mouse development. Mol. Cell. Biol. 35, 2910–2917 (2015).
Nakagawa, S. et al. The lncRNA Neat1 is required for corpus luteum formation and the establishment of pregnancy in a subpopulation of mice. Development 141, 4618–4627 (2014).
Standaert, L. et al. The long noncoding RNA Neat1 is required for mammary gland development and lactation. RNA 20, 1844–1849 (2014).
Jacks, T. et al. Tumor spectrum analysis in p53-mutant mice. Curr. Biol. 4, 1–7 (1994).
Vasioukhin, V. et al. The magical touch: genome targeting in epidermal stem cells induced by tamoxifen application to mouse skin. Proc. Natl. Acad. Sci. USA 96, 8551–8556 (1999).
Abel, E.L., Angel, J.M., Kiguchi, K. & DiGiovanni, J. Multi-stage chemical carcinogenesis in mouse skin: fundamentals and applications. Nat. Protoc. 4, 1350–1362 (2009).
Trapnell, C., Pachter, L. & Salzberg, S.L. TopHat: discovering splice junctions with RNA-seq. Bioinformatics 25, 1105–1111 (2009).
Anders, S., Pyl, P.T. & Huber, W. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169 (2015).
Harrow, J. et al. GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res. 22, 1760–1774 (2012).
Anders, S. & Huber, W. Differential expression analysis for sequence count data. Genome Biol. 11, R106 (2010).
Zhang, Y. et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9, R137 (2008).
Smoot, M.E., Ono, K., Ruscheinski, J., Wang, P.L. & Ideker, T. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics 27, 431–432 (2011).
Acknowledgements
We thank O. Van Goethem, G. Bervoets and S. Peeters for excellent technical assistance; E. Bonomi and M. Bugatti (Fondazione Beretta) for ISH analysis of human cancer samples; and S. Jackson for helpful comments on the manuscript and for imaging facility access. Imaging was done on a Nikon A1 confocal microscope acquired through a Hercules grant type 1 (AKUL/09/037) to W. Annaert. This work was supported by Interuniversitaire Attractiepolen (IUAP), an Institute for Science, Innovation and Technology (IWT) scholarship to C.A. and a Fund for Scientific Research Flanders (FWO) scholarship to L.S. W.V. is supported by Associazione Italiana per la Ricerca sul Cancro (AIRC, IG 15378) and by Ministero Salute (RF-2010-2315888).
Author information
Authors and Affiliations
Contributions
C.A., L.S. and J.B. designed and conducted experiments and acquired, analyzed and interpreted the data. M.L. performed DMBA and TPA treatments and monitored tumor development. A.V. and S.A. performed p53 ChIP-seq and RNA-seq experiments and data analysis. P.K. assessed DNA damage repair efficacy with bleomycin. B. Boeckx and D.L. analyzed expression data and constructed KM curves. E.R. and J.v.d.O. provided mouse and human pathology support. J.v.d.O. provided clinical samples. A.A.S., C.B. and E.L. designed research studies and contributed to interpretation of the data. G.L. and B. Beck provided reagents from mouse skin tumors. S.N. and T.H. provided Neat1 KO mice. W.V. performed the TMA. P.W.G.W. provided reagents and contributed to interpretation of the data. All authors read and edited the manuscript. J.-C.M. designed research studies and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–8 and Supplementary Tables 1–2 (PDF 18755 kb)
Rights and permissions
About this article
Cite this article
Adriaens, C., Standaert, L., Barra, J. et al. p53 induces formation of NEAT1 lncRNA-containing paraspeckles that modulate replication stress response and chemosensitivity. Nat Med 22, 861–868 (2016). https://doi.org/10.1038/nm.4135
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nm.4135
This article is cited by
-
Genome-scale pan-cancer interrogation of lncRNA dependencies using CasRx
Nature Methods (2024)
-
Combined absence of TRP53 target genes ZMAT3, PUMA and p21 cause a high incidence of cancer in mice
Cell Death & Differentiation (2024)
-
Effective methods for bulk RNA-seq deconvolution using scnRNA-seq transcriptomes
Genome Biology (2023)
-
Shell protein composition specified by the lncRNA NEAT1 domains dictates the formation of paraspeckles as distinct membraneless organelles
Nature Cell Biology (2023)
-
Targeting crosstalk of signaling pathways in cancer stem cells: a promising approach for development of novel anti-cancer therapeutics
Medical Oncology (2023)