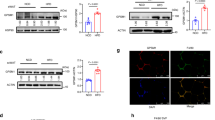
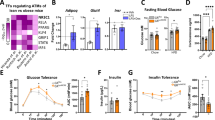
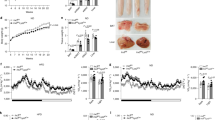
Abstract
Humans with obesity differ in their susceptibility to developing insulin resistance and type 2 diabetes (T2D). This variation may relate to the extent of adipose tissue (AT) inflammation that develops as their obesity progresses. The state of macrophage activation has a central role in determining the degree of AT inflammation and thus its dysfunction, and these states are driven by epigenomic alterations linked to gene expression. The underlying mechanisms that regulate these alterations, however, are poorly defined. Here we demonstrate that a co-repressor complex containing G protein pathway suppressor 2 (GPS2) crucially controls the macrophage epigenome during activation by metabolic stress. The study of AT from humans with and without obesity revealed correlations between reduced GPS2 expression in macrophages, elevated systemic and AT inflammation, and diabetic status. The causality of this relationship was confirmed by using macrophage-specific Gps2-knockout (KO) mice, in which inappropriate co-repressor complex function caused enhancer activation, pro-inflammatory gene expression and hypersensitivity toward metabolic-stress signals. By contrast, transplantation of GPS2-overexpressing bone marrow into two mouse models of obesity (ob/ob and diet-induced obesity) reduced inflammation and improved insulin sensitivity. Thus, our data reveal a potentially reversible disease mechanism that links co-repressor-dependent epigenomic alterations in macrophages to AT inflammation and the development of T2D.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Glass, C.K. & Olefsky, J.M. Inflammation and lipid signaling in the etiology of insulin resistance. Cell Metab. 15, 635–645 (2012).
Murray, P.J. et al. Macrophage activation and polarization: nomenclature and experimental guidelines. Immunity 41, 14–20 (2014).
Dalmas, E., Clément, K. & Guerre-Millo, M. Defining macrophage phenotype and function in adipose tissue. Trends Immunol. 32, 307–314 (2011).
Dalmas, E. et al. Irf5 deficiency in macrophages promotes beneficial adipose tissue expansion and insulin sensitivity during obesity. Nat. Med. 21, 610–618 (2015).
Kratz, M. et al. Metabolic dysfunction drives a mechanistically distinct proinflammatory phenotype in adipose tissue macrophages. Cell Metab. 20, 614–625 (2014).
Lumeng, C.N., Bodzin, J.L. & Saltiel, A.R. Obesity induces a phenotypic switch in adipose tissue macrophage polarization. J. Clin. Invest. 117, 175–184 (2007).
Nagareddy, P.R. et al. Adipose tissue macrophages promote myelopoiesis and monocytosis in obesity. Cell Metab. 19, 821–835 (2014).
Sun, K., Kusminski, C.M. & Scherer, P.E. Adipose tissue remodeling and obesity. J. Clin. Invest. 121, 2094–2101 (2011).
Tchernof, A. & Després, J.P. Pathophysiology of human visceral obesity: an update. Physiol. Rev. 93, 359–404 (2013).
Xu, X. et al. Obesity activates a program of lysosomal-dependent lipid metabolism in adipose tissue macrophages independently of classic activation. Cell Metab. 18, 816–830 (2013).
Kanda, H. et al. MCP-1 contributes to macrophage infiltration into adipose tissue, insulin resistance, and hepatic steatosis in obesity. J. Clin. Invest. 116, 1494–1505 (2006).
Mauer, J. et al. Signaling by IL-6 promotes alternative activation of macrophages to limit endotoxemia and obesity-associated resistance to insulin. Nat. Immunol. 15, 423–430 (2014).
Arner, E., Rydén, M. & Arner, P. Tumor necrosis factor alpha and regulation of adipose tissue. N. Engl. J. Med. 362, 1151–1153 (2010).
Gregor, M.F. & Hotamisligil, G.S. Inflammatory mechanisms in obesity. Annu. Rev. Immunol. 29, 415–445 (2011).
Gosselin, D. & Glass, C.K. Epigenomics of macrophages. Immunol. Rev. 262, 96–112 (2014).
Hah, N. et al. Inflammation-sensitive super enhancers form domains of coordinately regulated enhancer RNAs. Proc. Natl. Acad. Sci. USA 112, E297–E302 (2015).
Kaikkonen, M.U. et al. Remodeling of the enhancer landscape during macrophage activation is coupled to enhancer transcription. Mol. Cell 51, 310–325 (2013).
Link, V.M., Gosselin, D. & Glass, C.K. Mechanisms underlying the selection and function of macrophage-specific enhancers. Cold Spring Harb. Symp. Quant. Biol. 027367 (2015).
Glass, C.K. & Natoli, G. Molecular control of activation and priming in macrophages. Nat. Immunol. 17, 26–33 (2016).
Medzhitov, R. & Horng, T. Transcriptional control of the inflammatory response. Nat. Rev. Immunol. 9, 692–703 (2009).
Lawrence, T. & Natoli, G. Transcriptional regulation of macrophage polarization: enabling diversity with identity. Nat. Rev. Immunol. 11, 750–761 (2011).
Guenther, M.G. et al. A core SMRT corepressor complex containing HDAC3 and TBL1, a WD40-repeat protein linked to deafness. Genes Dev. 14, 1048–1057 (2000).
Guenther, M.G., Barak, O. & Lazar, M.A. The SMRT and N-CoR corepressors are activating cofactors for histone deacetylase 3. Mol. Cell. Biol. 21, 6091–6101 (2001).
Zhang, J., Kalkum, M., Chait, B.T. & Roeder, R.G. The N-CoR-HDAC3 nuclear receptor corepressor complex inhibits the JNK pathway through the integral subunit GPS2. Mol. Cell 9, 611–623 (2002).
Oberoi, J. et al. Structural basis for the assembly of the SMRT/NCoR core transcriptional repression machinery. Nat. Struct. Mol. Biol. 18, 177–184 (2011).
Barish, G.D. et al. The Bcl6-SMRT/NCoR cistrome represses inflammation to attenuate atherosclerosis. Cell Metab. 15, 554–562 (2012).
Ghisletti, S. et al. Cooperative NCoR/SMRT interactions establish a corepressor-based strategy for integration of inflammatory and anti-inflammatory signaling pathways. Genes Dev. 23, 681–693 (2009).
Glass, C.K. & Saijo, K. Nuclear receptor transrepression pathways that regulate inflammation in macrophages and T cells. Nat. Rev. Immunol. 10, 365–376 (2010).
Venteclef, N. et al. GPS2-dependent corepressor/SUMO pathways govern anti-inflammatory actions of LRH-1 and LXRbeta in the hepatic acute phase response. Genes Dev. 24, 381–395 (2010).
Toubal, A. et al. SMRT-GPS2 corepressor pathway dysregulation coincides with obesity-linked adipocyte inflammation. J. Clin. Invest. 123, 362–379 (2013).
Cardamone, M.D. et al. A protective strategy against hyperinflammatory responses requiring the nontranscriptional actions of GPS2. Mol. Cell 46, 91–104 (2012).
Li, P. et al. Adipocyte NCoR knockout decreases PPARγ phosphorylation and enhances PPARγ activity and insulin sensitivity. Cell 147, 815–826 (2011).
Li, P. et al. NCoR repression of LXRs restricts macrophage biosynthesis of insulin-sensitizing omega 3 fatty acids. Cell 155, 200–214 (2013).
Lavin, Y. et al. Tissue-resident macrophage enhancer landscapes are shaped by the local microenvironment. Cell 159, 1312–1326 (2014).
Mullican, S.E. et al. Histone deacetylase 3 is an epigenomic brake in macrophage alternative activation. Genes Dev. 25, 2480–2488 (2011).
Chen, X. et al. Requirement for the histone deacetylase Hdac3 for the inflammatory gene expression program in macrophages. Proc. Natl. Acad. Sci. USA 109, E2865–E2874 (2012).
Sun, Z. et al. Deacetylase-independent function of HDAC3 in transcription and metabolism requires nuclear receptor corepressor. Mol. Cell 52, 769–782 (2013).
Uhlenhaut, N.H. et al. Insights into negative regulation by the glucocorticoid receptor from genome-wide profiling of inflammatory cistromes. Mol. Cell 49, 158–171 (2013).
Cardamone, M.D. et al. GPS2/KDM4A pioneering activity regulates promoter-specific recruitment of PPARγ. Cell Rep. 8, 163–176 (2014).
Ahima, R.S. & Lazar, M.A. Physiology. The health risk of obesity—better metrics imperative. Science 341, 856–858 (2013).
Kang, S. et al. Identification of nuclear hormone receptor pathways causing insulin resistance by transcriptional and epigenomic analysis. Nat. Cell Biol. 17, 44–56 (2015).
Toubal, A., Treuter, E., Clément, K. & Venteclef, N. Genomic and epigenomic regulation of adipose tissue inflammation in obesity. Trends Endocrinol. Metab. 24, 625–634 (2013).
Stasevich, T.J. et al. Regulation of RNA polymerase II activation by histone acetylation in single living cells. Nature 516, 272–275 (2014).
Ito, A. et al. LXRs link metabolism to inflammation through Abca1-dependent regulation of membrane composition and TLR signaling. eLife 4, e08009 (2015).
Jonas, B.A. & Privalsky, M.L. SMRT and N-CoR corepressors are regulated by distinct kinase signaling pathways. J. Biol. Chem. 279, 54676–54686 (2004).
Hambleton, J., Weinstein, S.L., Lem, L. & DeFranco, A.L. Activation of c-Jun N-terminal kinase in bacterial lipopolysaccharide-stimulated macrophages. Proc. Natl. Acad. Sci. USA 93, 2774–2778 (1996).
Wolter, S. et al. c-Jun controls histone modifications, NF-kappaB recruitment, and RNA polymerase II function to activate the ccl2 gene. Mol. Cell. Biol. 28, 4407–4423 (2008).
Weiss, C. et al. JNK phosphorylation relieves HDAC3-dependent suppression of the transcriptional activity of c-Jun. EMBO J. 22, 3686–3695 (2003).
Lee, S.K., Kim, J.H., Lee, Y.C., Cheong, J. & Lee, J.W. Silencing mediator of retinoic acid and thyroid hormone receptors, as a novel transcriptional corepressor molecule of activating protein-1, nuclear factor-κβ, and serum response factor. J. Biol. Chem. 275, 12470–12474 (2000).
Ogawa, S. et al. A nuclear receptor corepressor transcriptional checkpoint controlling activator protein 1-dependent gene networks required for macrophage activation. Proc. Natl. Acad. Sci. USA 101, 14461–14466 (2004).
Garber, M. et al. A high-throughput chromatin immunoprecipitation approach reveals principles of dynamic gene regulation in mammals. Mol. Cell 47, 810–822 (2012).
Siersbæk, R. et al. Transcription factor cooperativity in early adipogenic hotspots and super-enhancers. Cell Rep. 7, 1443–1455 (2014).
Carvalho, B.S. & Irizarry, R.A. A framework for oligonucleotide microarray preprocessing. Bioinformatics 26, 2363–2367 (2010).
Ritchie, M.E. et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47 (2015).
Wickham, H. ggplot2: Elegant Graphics for Data Analysis (Springer, 2009).
Behrens, A. et al. Impaired postnatal hepatocyte proliferation and liver regeneration in mice lacking c-jun in the liver. EMBO J. 21, 1782–1790 (2002).
Fogg, D.K. et al. A clonogenic bone marrow progenitor specific for macrophages and dendritic cells. Science 311, 83–87 (2006).
Schulz, C. et al. A lineage of myeloid cells independent of Myb and hematopoietic stem cells. Science 336, 86–90 (2012).
Prieur, X. et al. Differential lipid partitioning between adipocytes and tissue macrophages modulates macrophage lipotoxicity and M2/M1 polarization in obese mice. Diabetes 60, 797–809 (2011).
Núñez, V. et al. Retinoid X receptor alpha controls innate inflammatory responses through the up-regulation of chemokine expression. Proc. Natl. Acad. Sci. USA 107, 10626–10631 (2010).
Schmidt, D. et al. ChIP-seq: using high-throughput sequencing to discover protein-DNA interactions. Methods 48, 240–248 (2009).
Heinz, S. et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38, 576–589 (2010).
Robinson, M.D., McCarthy, D.J. & Smyth, G.K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010).
McCarthy, D.J., Chen, Y. & Smyth, G.K. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res. 40, 4288–4297 (2012).
Ran, F.A. et al. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 8, 2281–2308 (2013).
Gautier, E.L. et al. HDL and Glut1 inhibition reverse a hypermetabolic state in mouse models of myeloproliferative disorders. J. Exp. Med. 210, 339–353 (2013).
Hajduch, E., Darakhshan, F. & Hundal, H.S. Fructose uptake in rat adipocytes: GLUT5 expression and the effects of streptozotocin-induced diabetes. Diabetologia 41, 821–828 (1998).
Acknowledgements
We acknowledge Ozgene Pty, Ltd. for generating floxed GPS2 mice. We acknowledge V. Benes and the team at the EMBL Genomics Core Facility for ChIP-seq library preparation and sequencing. We thank S. Mandrup (University of Southern Denmark), K. de Bosscher (Ghent University), J. Taipale (Karolinska Institutet), and F. Zhang (Broad Institute) for providing plasmids, and T. Jakobsson for his advice in establishing the CRISPR–Cas9 system. We also thank F. Fagerström-Billai and co-workers at the BEA Core Facility (Karolinska Institutet) for microarray preparation and analysis. For assistance with pathological analysis and FACS sorting, we thank R. Kuiper, T. Schröder, and Å.-L. Dackland (Department of Laboratory Medicine, Karolinska Institutet). We thank R. Chiche, J. Cady, and S. Gueroult (Geoffroy Saint-Hilaire Clinic Paris) for human AT sampling, and J.-L. Nguewa for his help in PHRC Glucostress. This work was supported by grants from the Swedish Research Council (Vetenskapsrådet, E.T.); the Swedish Cancer Society (Cancerfonden, E.T.); the Swedish Diabetes Foundation (Diabetesfonden, E.T.); the Novo Nordisk Foundation (Novo Nordisk Fonden, E.T.); the Center for Biosciences (former CB, now CIMED) at Karolinska Institutet (E.T.); the French National Agency of Research (CONRAD, N.V.; ANR CE12 2014, FATMAC, T.B.); Region Ile de France (CORDDIM, N.V.); Paris city (EMERGENCE, N.V.); the French Foundation for Medical Research (Equipe FRM DEQ20140329504, N.V. and F.F.); a French government grant managed by the National Agency of Research (program 'Investments for the Future' reference ANR-10-IAH, ICAN MetaMACS, N.V. and F.F.); Assistance Publique des Hôpitaux de Paris (APHP), Programs of Clinical Investigation (CRC Fibrota AOO759-32 to J.A.-W.; and PHRC Glucostress P081122 to J.-F.G.). A.T. and K.D. received a doctoral fellowship from the 'Ministère de la Recherche et de l'Enseignement supérieur'. Z.H. received a doctoral fellowship from the China Scholarship Council (CRC), and N.L. received a doctoral faculty grant from the Karolinska Institutet (KID funding).
Author information
Authors and Affiliations
Contributions
R.F., A.T. and S.G. share the first authorship. K.D. and Z.H. share the second authorship. R.F. and A.T. performed the majority of in vivo experiments, with contributions from K.D., I.H., R.B., and F.A., and analyzed data. S.G. performed the genomic and ChIP-seq studies, with major contribution from Z.H., and A.D. and R.F. analyzed genomic data. N.L. contributed to the in vitro studies. F.F., J.-F.G., J.A.-W., A.S., A.T., P.A., and N.V. collected, analyzed, and interpreted the human data. T.L. provided Jun KO mice and analyzed data. R.F., N.V., and E.T. conceived the study, interpreted data, and wrote the manuscript. N.V. and E.T. supervised the study and share the corresponding authorship.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–8 and Supplementary Tables 1–6 (PDF 14535 kb)
Rights and permissions
About this article
Cite this article
Fan, R., Toubal, A., Goñi, S. et al. Loss of the co-repressor GPS2 sensitizes macrophage activation upon metabolic stress induced by obesity and type 2 diabetes. Nat Med 22, 780–791 (2016). https://doi.org/10.1038/nm.4114
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nm.4114
This article is cited by
-
Macrophage function in adipose tissue homeostasis and metabolic inflammation
Nature Immunology (2023)
-
Uncurtaining the pivotal role of ABC transporters in diabetes mellitus
Environmental Science and Pollution Research (2021)
-
Prospective analyses of white adipose tissue gene expression in relation to long-term body weight changes
International Journal of Obesity (2020)
-
Dopaminergic Pathways in Obesity-Associated Inflammation
Journal of Neuroimmune Pharmacology (2020)
-
Transcriptional control of macrophage polarisation in type 2 diabetes
Seminars in Immunopathology (2019)