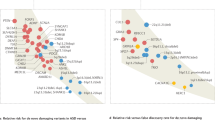
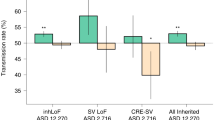
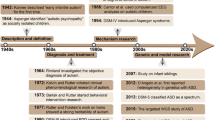
Abstract
Progress in understanding the genetic etiology of autism spectrum disorders (ASD) has fueled remarkable advances in our understanding of its potential neurobiological mechanisms. Yet, at the same time, these findings highlight extraordinary causal diversity and complexity at many levels ranging from molecules to circuits and emphasize the gaps in our current knowledge. Here we review current understanding of the genetic architecture of ASD and integrate genetic evidence, neuropathology and studies in model systems with how they inform mechanistic models of ASD pathophysiology. Despite the challenges, these advances provide a solid foundation for the development of rational, targeted molecular therapies.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Altshuler, D., Daly, M.J. & Lander, E.S. Genetic mapping in human disease. Science 322, 881–888 (2008).
Gilissen, C. et al. Genome sequencing identifies major causes of severe intellectual disability. Nature 511, 344–347 (2014).
Lee, H. et al. Clinical exome sequencing for genetic identification of rare Mendelian disorders. J. Am. Med. Assoc. 312, 1880–1887 (2014).
Psychiatric GWAS Consortium Bipolar Disorder Working Group. Large-scale genome-wide association analysis of bipolar disorder identifies a new susceptibility locus near ODZ4. Nat. Genet. 43, 977–983 (2011).
Lambert, J.C. et al. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer's disease. Nat. Genet. 45, 1452–1458 (2013).
Schizophrenia Working Group of the Psychiatric Genomics Consortium. Biological insights from 108 schizophrenia-associated genetic loci. Nature 511, 421–427 (2014).
Hindorff, L.A. et al. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc. Natl. Acad. Sci. USA 106, 9362–9367 (2009).
Geschwind, D.H. Autism: many genes, common pathways? Cell 135, 391–395 (2008).
Geschwind, D.H. Genetics of autism spectrum disorders. Trends Cogn. Sci. 15, 409–416 (2011).
Chen, J.A., Peñagarikano, O., Belgard, T.G., Swarup, V. & Geschwind, D.H. The emerging picture of autism spectrum disorder: genetics and pathology. Annu. Rev. Pathol. 10, 111–144 (2015).
Geschwind, D.H. & Levitt, P. Autism spectrum disorders: developmental disconnection syndromes. Curr. Opin. Neurobiol. 17, 103–111 (2007).
American Psychiatric Association. Diagnostic and Statistical Manual of Mental Disorders: DSM-5 5th edn. (American Psychiatric Association, Arlington, Viginia, USA, 2013).
Autism and Developmental Disabilities Monitoring Network Surveillance Year 2010 Principal Investigators. Prevalence of autism spectrum disorder among children aged 8 years—Autism and Developmental Disabilities Monitoring Network, 11 sites, United States, 2010. MMWR Surveill. Summ. 63, 1–21 (2014).
Werling, D.M. & Geschwind, D.H. Understanding sex bias in autism spectrum disorder. Proc. Natl. Acad. Sci. USA 110, 4868–4869 (2013).
Robinson, E.B., Lichtenstein, P., Anckarsäter, H., Happé, F. & Ronald, A. Examining and interpreting the female protective effect against autistic behavior. Proc. Natl. Acad. Sci. USA 110, 5258–5262 (2013).
Geschwind, D.H. Advances in autism. Annu. Rev. Med. 60, 367–380 (2009).
Volkmar, F.R. & McPartland, J.C. From Kanner to DSM-5: autism as an evolving diagnostic concept. Annu. Rev. Clin. Psychol. 10, 193–212 (2014).
Geschwind, D.H. & State, M.W. Gene hunting in autism spectrum disorder: on the path to precision medicine. Lancet Neurol. 14, 1109–1120 (2015).
Rogers, S.J. et al. Autism treatment in the first year of life: a pilot study of infant start, a parent-implemented intervention for symptomatic infants. J. Autism Dev. Disord. 44, 2981–2995 (2014).
Kasari, C., Shire, S., Factor, R. & McCracken, C. Psychosocial treatments for individuals with autism spectrum disorder across the lifespan: new developments and underlying mechanisms. Curr. Psychiatry Rep. 16, 512 (2014).
Jones, W. & Klin, A. Attention to eyes is present but in decline in 2- to 6-month-old infants later diagnosed with autism. Nature 504, 427–431 (2013).
Jeste, S.S., Frohlich, J. & Loo, S.K. Electrophysiological biomarkers of diagnosis and outcome in neurodevelopmental disorders. Curr. Opin. Neurol. 28, 110–116 (2015).
Zafeiriou, D.I., Ververi, A., Dafoulis, V., Kalyva, E. & Vargiami, E. Autism spectrum disorders: the quest for genetic syndromes. Am. J. Med. Genet. B. Neuropsychiatr. Genet. 162B, 327–366 (2013).
Miles, J.H. Autism spectrum disorders—a genetics review. Genet. Med. 13, 278–294 (2011).
Betancur, C. Etiological heterogeneity in autism spectrum disorders: more than 100 genetic and genomic disorders and still counting. Brain Res. 1380, 42–77 (2011).
Gillberg, C. & Coleman, M. The Biology of the Autistic Syndromes (Mac Keith Press, London, 1992).
Folstein, S. & Rutter, M. Genetic influences and infantile autism. Nature 265, 726–728 (1977).
Hallmayer, J. et al. Genetic heritability and shared environmental factors among twin pairs with autism. Arch. Gen. Psychiatry 68, 1095–1102 (2011).
Sandin, S. et al. The familial risk of autism. J. Am. Med. Assoc. 311, 1770–1777 (2014).
Bailey, A. et al. Autism as a strongly genetic disorder: evidence from a British twin study. Psychol. Med. 25, 63–77 (1995).
Lichtenstein, P., Carlström, E., Råstam, M., Gillberg, C. & Anckarsäter, H. The genetics of autism spectrum disorders and related neuropsychiatric disorders in childhood. Am. J. Psychiatry 167, 1357–1363 (2010).
Ronald, A. & Hoekstra, R.A. Autism spectrum disorders and autistic traits: a decade of new twin studies. Am. J. Med. Genet. B. Neuropsychiatr. Genet. 156B, 255–274 (2011).
Sebat, J. et al. Strong association of de novo copy-number mutations with autism. Science 316, 445–449 (2007).
Szatmari, P. et al. Mapping autism risk loci using genetic linkage and chromosomal rearrangements. Nat. Genet. 39, 319–328 (2007).
Marshall, C.R. et al. Structural variation of chromosomes in autism spectrum disorder. Am. J. Hum. Genet. 82, 477–488 (2008).
Bucan, M. et al. Genome-wide analyses of exonic copy-number variants in a family-based study point to novel autism susceptibility genes. PLoS Genet. 5, e1000536 (2009).
Glessner, J.T. et al. Autism genome-wide copy-number variation reveals ubiquitin and neuronal genes. Nature 459, 569–573 (2009).
Itsara, A. et al. De novo rates and selection of large copy-number variation. Genome Res. 20, 1469–1481 (2010).
Pinto, D. et al. Functional impact of global, rare copy-number variation in autism spectrum disorders. Nature 466, 368–372 (2010).
Levy, D. et al. Rare de novo and transmitted copy-number variation in autistic spectrum disorders. Neuron 70, 886–897 (2011).
Griswold, A.J. et al. Evaluation of copy-number variations reveals novel candidate genes in autism spectrum disorder–associated pathways. Hum. Mol. Genet. 21, 3513–3523 (2012).
Neale, B.M. et al. Patterns and rates of exonic de novo mutations in autism spectrum disorders. Nature 485, 242–245 (2012).
De Rubeis, S. et al. Synaptic, transcriptional and chromatin genes disrupted in autism. Nature 515, 209–215 (2014).
Iossifov, I. et al. The contribution of de novo coding mutations to autism spectrum disorder. Nature 515, 216–221 (2014).
Iossifov, I. et al. De novo gene disruptions in children on the autistic spectrum. Neuron 74, 285–299 (2012).
O'Roak, B.J. et al. Sporadic autism exomes reveal a highly interconnected protein network of de novo mutations. Nature 485, 246–250 (2012).
Sanders, S.J. et al. De novo mutations revealed by whole-exome sequencing are strongly associated with autism. Nature 485, 237–241 (2012).
Gilman, S.R. et al. Rare de novo variants associated with autism implicate a large functional network of genes involved in formation and function of synapses. Neuron 70, 898–907 (2011).
Sanders, S.J. et al. Multiple recurrent de novo CNVs, including duplications of the 7q11.23 Williams syndrome region, are strongly associated with autism. Neuron 70, 863–885 (2011).
Gratten, J., Visscher, P.M., Mowry, B.J. & Wray, N.R. Interpreting the role of de novo protein-coding mutations in neuropsychiatric disease. Nat. Genet. 45, 234–238 (2013).
O'Roak, B.J. et al. Multiplex targeted sequencing identifies recurrently mutated genes in autism spectrum disorders. Science 338, 1619–1622 (2012).
Morrow, E.M. et al. Identifying autism loci and genes by tracing recent shared ancestry. Science 321, 218–223 (2008).
Strauss, K.A. et al. Recessive symptomatic focal epilepsy and mutant contactin-associated protein-like 2. N. Engl. J. Med. 354, 1370–1377 (2006).
Lim, E.T. et al. Rare complete knockouts in humans: population distribution and significant role in autism spectrum disorders. Neuron 77, 235–242 (2013).
Yu, T.W. et al. Using whole-exome sequencing to identify inherited causes of autism. Neuron 77, 259–273 (2013).
Lee, S.H. et al. Estimating the proportion of variation in susceptibility to schizophrenia, captured by common SNPs. Nat. Genet. 44, 247–250 (2012).
Gaugler, T. et al. Most genetic risk for autism resides with common variation. Nat. Genet. 46, 881–885 (2014).
Bulik-Sullivan, B.K. et al. LD score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat. Genet. 47, 291–295 (2015).
Wray, N.R. & Visscher, P.M. Quantitative genetics of disease traits. J. Anim. Breed. Genet. 132, 198–203 (2015).
O'Roak, B.J. et al. Exome sequencing in sporadic autism spectrum disorders identifies severe de novo mutations. Nat. Genet. 43, 585–589 (2011).
Schaaf, C.P. et al. Oligogenic heterozygosity in individuals with high-functioning autism spectrum disorders. Hum. Mol. Genet. 20, 3366–3375 (2011).
Zhao, X. et al. A unified genetic theory for sporadic and inherited autism. Proc. Natl. Acad. Sci. USA 104, 12831–12836 (2007).
Ronemus, M., Iossifov, I., Levy, D. & Wigler, M. The role of de novo mutations in the genetics of autism spectrum disorders. Nat. Rev. Genet. 15, 133–141 (2014).
Virkud, Y.V., Todd, R.D., Abbacchi, A.M., Zhang, Y. & Constantino, J.N. Familial aggregation of quantitative autistic traits in multiplex versus simplex autism. Am. J. Med. Genet. B. Neuropsychiatr. Genet. 150B, 328–334 (2009).
Lyall, K. et al. Parental social responsiveness and risk of autism spectrum disorder in offspring. JAMA Psychiatry 71, 936–942 (2014).
Lowe, J.K., Werling, D.M., Constantino, J.N., Cantor, R.M. & Geschwind, D.H. Social responsiveness, an autism endophenotype: genome-wide significant linkage to two regions on chromosome 8. Am. J. Psychiatry 172, 266–275 (2015).
Lee, S.H. et al. Genetic relationship between five psychiatric disorders estimated from genome-wide SNPs. Nat. Genet. 45, 984–994 (2013).
Klei, L. et al. Common genetic variants, acting additively, are a major source of risk for autism. Mol. Autism 3, 9 (2012).
Maier, R. et al. Joint analysis of psychiatric disorders increases accuracy of risk prediction for schizophrenia, bipolar disorder and major depressive disorder. Am. J. Hum. Genet. 96, 283–294 (2015).
Anney, R. et al. Individual common variants exert weak effects on the risk for autism spectrum disorders. Hum. Mol. Genet. 21, 4781–4792 (2012).
Gratten, J., Wray, N.R., Keller, M.C. & Visscher, P.M. Large-scale genomics unveils the genetic architecture of psychiatric disorders. Nat. Neurosci. 17, 782–790 (2014).
Geschwind, D.H. & Flint, J. Genetics and genomics of psychiatric disease. Science 349, 1489–1494 (2015).
Krumm, N. et al. Excess of rare, inherited truncating mutations in autism. Nat. Genet. 47, 582–588 (2015).
Pinto, D. et al. Convergence of genes and cellular pathways dysregulated in autism spectrum disorders. Am. J. Hum. Genet. 94, 677–694 (2014).
Vinkhuyzen, A.A., Wray, N.R., Yang, J., Goddard, M.E. & Visscher, P.M. Estimation and partition of heritability in human populations using whole-genome analysis methods. Annu. Rev. Genet. 47, 75–95 (2013).
Sham, P.C. & Purcell, S.M. Statistical power and significance testing in large-scale genetic studies. Nat. Rev. Genet. 15, 335–346 (2014).
Stessman, H.A., Bernier, R. & Eichler, E.E. A genotype-first approach to defining the subtypes of a complex disease. Cell 156, 872–877 (2014).
Ji, J. et al. DYRK1A haploinsufficiency causes a new recognizable syndrome with microcephaly, intellectual disability, speech impairment and distinct facies. Eur. J. Hum. Genet. 23, 1473–1481 (2015).
Bronicki, L.M. et al. Ten new cases further delineate the syndromic intellectual disability phenotype caused by mutations in DYRK1A. Eur. J. Hum. Genet. 23, 1482–1487 (2015).
Bernier, R. et al. Disruptive CHD8 mutations define a subtype of autism early in development. Cell 158, 263–276 (2014).
Jablensky, A. Schizophrenia or schizophrenias? The challenge of genetic parsing of a complex disorder. Am. J. Psychiatry 172, 105–107 (2015).
Skafidas, E. et al. Predicting the diagnosis of autism spectrum disorder using gene pathway analysis. Mol. Psychiatry 19, 504–510 (2014).
Arnedo, J. et al. Uncovering the hidden risk architecture of the schizophrenias: confirmation in three independent genome-wide association studies. Am. J. Psychiatry 172, 139–153 (2015).
Robinson, E.B. et al. Response to 'Predicting the diagnosis of autism spectrum disorder using gene pathway analysis'. Mol. Psychiatry 19, 859–861 (2014).
Belgard, T.G., Jankovic, I., Lowe, J.K. & Geschwind, D.H. Population structure confounds autism genetic classifier. Mol. Psychiatry 19, 405–407 (2014).
Noh, H.J. et al. Network topologies and convergent etiologies arising from deletions and duplications observed in individuals with autism. PLoS Genet. 9, e1003523 (2013).
Willsey, A.J. et al. Coexpression networks implicate human midfetal deep cortical projection neurons in the pathogenesis of autism. Cell 155, 997–1007 (2013).
Parikshak, N.N. et al. Integrative functional genomic analyses implicate specific molecular pathways and circuits in autism. Cell 155, 1008–1021 (2013).
Voineagu, I. et al. Transcriptomic analysis of autistic brain reveals convergent molecular pathology. Nature 474, 380–384 (2011).
Abrahams, B.S. & Geschwind, D.H. Advances in autism genetics: on the threshold of a new neurobiology. Nat. Rev. Genet. 9, 341–355 (2008).
Pankevich, D.E., Wizemann, T.M. & Altevogt, B.M. Improving the Utility and Translation of Animal Models for Nervous System Disorders: Workshop Summary (The National Academies Press, Washington, D.C., 2013).
Dolmetsch, R. & Geschwind, D.H. The human brain in a dish: the promise of iPSC-derived neurons. Cell 145, 831–834 (2011).
Silverman, J.L., Yang, M., Lord, C. & Crawley, J.N. Behavioral phenotyping assays for mouse models of autism. Nat. Rev. Neurosci. 11, 490–502 (2010).
Watson, K.K. & Platt, M.L. Of mice and monkeys: using nonhuman primate models to bridge mouse- and human-based investigations of autism spectrum disorders. J. Neurodev. Disord. 4, 21 (2012).
McCammon, J.M. & Sive, H. Addressing the genetics of human mental health disorders in model organisms. Annu. Rev. Genomics Hum. Genet. 16, 173–197 (2015).
Tian, Y. et al. Alteration in basal and depolarization-induced transcriptional network in iPSC-derived neurons from Timothy syndrome. Genome Med. 6, 75 (2014).
Fogel, B.L. et al. RBFOX1 regulates both splicing and transcriptional networks in human neuronal development. Hum. Mol. Genet. 21, 4171–4186 (2012).
Sugathan, A. et al. CHD8 regulates neurodevelopmental pathways associated with autism spectrum disorder in neural progenitors. Proc. Natl. Acad. Sci. USA 111, E4468–E4477 (2014).
Paşca, S.P. et al. Using iPSC-derived neurons to uncover cellular phenotypes associated with Timothy syndrome. Nat. Med. 17, 1657–1662 (2011).
Marchetto, M.C. et al. A model for neural development and treatment of Rett syndrome using human induced pluripotent stem cells. Cell 143, 527–539 (2010).
Li, Y. et al. Global transcriptional and translational repression in human embryonic stem cell–derived Rett syndrome neurons. Cell Stem Cell 13, 446–458 (2013).
Shcheglovitov, A. et al. SHANK3 and IGF1 restore synaptic deficits in neurons from 22q13 deletion syndrome patients. Nature 503, 267–271 (2013).
Mariani, J. et al. FOXG1-dependent dysregulation of GABA-glutamate neuron differentiation in autism spectrum disorders. Cell 162, 375–390 (2015).
Casanova, M.F. et al. Minicolumnar abnormalities in autism. Acta Neuropathol. 112, 287–303 (2006).
Stoner, R. et al. Patches of disorganization in the neocortex of children with autism. N. Engl. J. Med. 370, 1209–1219 (2014).
Redcay, E. & Courchesne, E. When is the brain enlarged in autism? A meta-analysis of all brain size reports. Biol. Psychiatry 58, 1–9 (2005).
Magri, L. et al. Sustained activation of mTOR pathway in embryonic neural stem cells leads to development of tuberous sclerosis complex–associated lesions. Cell Stem Cell 9, 447–462 (2011).
La Fata, G. et al. FMRP regulates multipolar to bipolar transition affecting neuronal migration and cortical circuitry. Nat. Neurosci. 17, 1693–1700 (2014).
Peñagarikano, O. et al. Absence of Cntnap2 leads to epilepsy, neuronal migration abnormalities and core autism-related deficits. Cell 147, 235–246 (2011).
Feng, W. et al. The chromatin remodeler CHD7 regulates adult neurogenesis via activation of SoxC transcription factors. Cell Stem Cell 13, 62–72 (2013).
Zhou, J. et al. Tsc1-mutant neural stem and progenitor cells exhibit migration deficits and give rise to subependymal lesions in the lateral ventricle. Genes Dev. 25, 1595–1600 (2011).
Sarbassov, D.D., Ali, S.M. & Sabatini, D.M. Growing roles for the mTOR pathway. Curr. Opin. Cell Biol. 17, 596–603 (2005).
Lipton, J.O. & Sahin, M. The neurology of mTOR. Neuron 84, 275–291 (2014).
Kwon, C.H. et al. Pten regulates neuronal arborization and social interaction in mice. Neuron 50, 377–388 (2006).
Krumm, N., O'Roak, B.J., Shendure, J. & Eichler, E.E. A de novo convergence of autism genetics and molecular neuroscience. Trends Neurosci. 37, 95–105 (2014).
Hansen, D.V., Rubenstein, J.L. & Kriegstein, A.R. Deriving excitatory neurons of the neocortex from pluripotent stem cells. Neuron 70, 645–660 (2011).
Tiberi, L., Vanderhaeghen, P. & van den Ameele, J. Cortical neurogenesis and morphogens: diversity of cues, sources and functions. Curr. Opin. Cell Biol. 24, 269–276 (2012).
Lijam, N. et al. Social interaction and sensorimotor-gating abnormalities in mice lacking Dvl1. Cell 90, 895–905 (1997).
Sowers, L.P. et al. Disruption of the noncanonical Wnt gene PRICKLE2 leads to autism-like behaviors with evidence for hippocampal synaptic dysfunction. Mol. Psychiatry 18, 1077–1089 (2013).
Fang, W.Q. et al. Overproduction of upper-layer neurons in the neocortex leads to autism-like features in mice. Cell Reports 9, 1635–1643 (2014).
Long, J.M., LaPorte, P., Paylor, R. & Wynshaw-Boris, A. Expanded characterization of the social-interaction abnormalities in mice lacking Dvl1. Genes Brain Behav. 3, 51–62 (2004).
Bedogni, F. et al. Tbr1 regulates regional and laminar identity of postmitotic neurons in developing neocortex. Proc. Natl. Acad. Sci. USA 107, 13129–13134 (2010).
Sakamoto, I. et al. A novel β-catenin–binding protein inhibits β-catenin–dependent Tcf activation and axis formation. J. Biol. Chem. 275, 32871–32878 (2000).
Schuettengruber, B., Martinez, A.M., Iovino, N. & Cavalli, G. Trithorax group proteins: switching genes on and keeping them active. Nat. Rev. Mol. Cell Biol. 12, 799–814 (2011).
Nozawa, R.S. et al. Human POGZ modulates dissociation of HP1-α from mitotic chromosome arms through Aurora B activation. Nat. Cell Biol. 12, 719–727 (2010).
Chen, T. & Dent, S.Y. Chromatin modifiers and remodelers: regulators of cellular differentiation. Nat. Rev. Genet. 15, 93–106 (2014).
Ronan, J.L., Wu, W. & Crabtree, G.R. From neural development to cognition: unexpected roles for chromatin. Nat. Rev. Genet. 14, 347–359 (2013).
Hevner, R.F. et al. Tbr1 regulates differentiation of the preplate and layer 6. Neuron 29, 353–366 (2001).
Sierra, J., Yoshida, T., Joazeiro, C.A. & Jones, K.A. The APC tumor suppressor counteracts β-catenin activation and H3K4 methylation at Wnt target genes. Genes Dev. 20, 586–600 (2006).
Stein, J.L. et al. A quantitative framework to evaluate modeling of cortical development by neural stem cells. Neuron 83, 69–86 (2014).
Miller, J.A. et al. Transcriptional landscape of the prenatal human brain. Nature 508, 199–206 (2014).
Tuoc, T.C. et al. Chromatin regulation by BAF170 controls cerebral cortical size and thickness. Dev. Cell 25, 256–269 (2013).
Cotney, J. et al. The autism-associated chromatin modifier CHD8 regulates other autism risk genes during human neurodevelopment. Nat. Commun. 6, 6404 (2015).
Nishiyama, M., Skoultchi, A.I. & Nakayama, K.I. Histone H1 recruitment by CHD8 is essential for suppression of the Wnt–β-catenin signaling pathway. Mol. Cell. Biol. 32, 501–512 (2012).
Vaags, A.K. et al. Rare deletions at the neurexin 3 locus in autism spectrum disorder. Am. J. Hum. Genet. 90, 133–141 (2012).
Gauthier, J. et al. Truncating mutations in NRXN2 and NRXN1 in autism spectrum disorders and schizophrenia. Hum. Genet. 130, 563–573 (2011).
Jamain, S. et al. Mutations of the X-linked genes encoding neuroligins NLGN3 and NLGN4 are associated with autism. Nat. Genet. 34, 27–29 (2003).
Berkel, S. et al. Mutations in the SHANK2 synaptic scaffolding gene in autism spectrum disorder and mental retardation. Nat. Genet. 42, 489–491 (2010).
Durand, C.M. et al. Mutations in the gene encoding the synaptic scaffolding protein SHANK3 are associated with autism spectrum disorders. Nat. Genet. 39, 25–27 (2007).
Piton, A. et al. Analysis of the effects of rare variants on splicing identifies alterations in GABAA receptor genes in autism spectrum disorder individuals. Eur. J. Hum. Genet. 21, 749–756 (2013).
Lionel, A.C. et al. Rare exonic deletions implicate the synaptic organizer gephyrin (GPHN) in risk for autism, schizophrenia and seizures. Hum. Mol. Genet. 22, 2055–2066 (2013).
Fassio, A. et al. SYN1 loss-of-function mutations in autism and partial epilepsy cause impaired synaptic function. Hum. Mol. Genet. 20, 2297–2307 (2011).
Corradi, A. et al. SYN2 is an autism predisposing gene: loss-of-function mutations alter synaptic vesicle cycling and axon outgrowth. Hum. Mol. Genet. 23, 90–103 (2014).
Tang, G. et al. Loss of mTOR-dependent macroautophagy causes autistic-like synaptic pruning deficits. Neuron 83, 1131–1143 (2014).
Hutsler, J.J. & Zhang, H. Increased dendritic spine densities on cortical projection neurons in autism spectrum disorders. Brain Res. 1309, 83–94 (2010).
Blatt, G.J. et al. Density and distribution of hippocampal neurotransmitter receptors in autism: an autoradiographic study. J. Autism Dev. Disord. 31, 537–543 (2001).
Oblak, A., Gibbs, T.T. & Blatt, G.J. Decreased GABAA receptors and benzodiazepine-binding sites in the anterior cingulate cortex in autism. Autism Res. 2, 205–219 (2009).
Oblak, A.L., Gibbs, T.T. & Blatt, G.J. Decreased GABAB receptors in the cingulate cortex and fusiform gyrus in autism. J. Neurochem. 114, 1414–1423 (2010).
Fatemi, S.H. et al. mRNA and protein levels for GABAA-α4, -α5, -β1 and GABABR1 receptors are altered in brains from subjects with autism. J. Autism Dev. Disord. 40, 743–750 (2010).
Yip, J., Soghomonian, J.J. & Blatt, G.J. Increased GAD67 mRNA expression in cerebellar interneurons in autism: implications for Purkinje cell dysfunction. J. Neurosci. Res. 86, 525–530 (2008).
Yip, J., Soghomonian, J.J. & Blatt, G.J. Decreased GAD65 mRNA levels in select subpopulations of neurons in the cerebellar dentate nuclei in autism: an in situ hybridization study. Autism Res. 2, 50–59 (2009).
Yip, J., Soghomonian, J.J. & Blatt, G.J. Decreased GAD67 mRNA levels in cerebellar Purkinje cells in autism: pathophysiological implications. Acta Neuropathol. 113, 559–568 (2007).
Fatemi, S.H. et al. Glutamic acid decarboxylase 65 and 67 kDa proteins are reduced in autistic parietal and cerebellar cortices. Biol. Psychiatry 52, 805–810 (2002).
Südhof, T.C. Neuroligins and neurexins link synaptic function to cognitive disease. Nature 455, 903–911 (2008).
Citri, A. & Malenka, R.C. Synaptic plasticity: multiple forms, functions and mechanisms. Neuropsychopharmacology 33, 18–41 (2008).
Mayford, M., Siegelbaum, S.A. & Kandel, E.R. Synapses and memory storage. Cold Spring Harb. Perspect. Biol. 4, a005751 (2012).
Yizhar, O. et al. Neocortical excitation-inhibition balance in information processing and social dysfunction. Nature 477, 171–178 (2011).
Palop, J.J. et al. Aberrant excitatory neuronal activity and compensatory remodeling of inhibitory hippocampal circuits in mouse models of Alzheimer's disease. Neuron 55, 697–711 (2007).
Fritschy, J.M. Epilepsy, E/I balance and GABAA receptor plasticity. Front. Mol. Neurosci. 1, 5 (2008).
Kehrer, C., Maziashvili, N., Dugladze, T. & Gloveli, T. Altered excitatory-inhibitory balance in the NMDA-hypofunction model of schizophrenia. Front. Mol. Neurosci. 1, 6 (2008).
van de Lagemaat, L.N. et al. Age-related decreased inhibitory versus excitatory gene expression in the adult autistic brain. Front. Neurosci. 8, 394 (2014).
Rojas, D.C., Singel, D., Steinmetz, S., Hepburn, S. & Brown, M.S. Decreased left perisylvian GABA concentration in children with autism and unaffected siblings. Neuroimage 86, 28–34 (2014).
Bejjani, A. et al. Elevated glutamatergic compounds in pregenual anterior cingulate in pediatric autism spectrum disorder demonstrated by 1H MRS and 1H MRSI. PLoS One 7, e38786 (2012).
Greer, P.L. & Greenberg, M.E. From synapse to nucleus: calcium-dependent gene transcription in the control of synapse development and function. Neuron 59, 846–860 (2008).
Buffington, S.A., Huang, W. & Costa-Mattioli, M. Translational control in synaptic plasticity and cognitive dysfunction. Annu. Rev. Neurosci. 37, 17–38 (2014).
Amir, R.E. et al. Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG–binding protein 2. Nat. Genet. 23, 185–188 (1999).
Splawski, I. et al. CaV1.2 calcium channel dysfunction causes a multisystem disorder, including arrhythmia and autism. Cell 119, 19–31 (2004).
Ebert, D.H. & Greenberg, M.E. Activity-dependent neuronal signaling and autism spectrum disorder. Nature 493, 327–337 (2013).
Nicholls, R.D. & Knepper, J.L. Genome organization, function and imprinting in Prader-Willi and Angelman syndromes. Annu. Rev. Genomics Hum. Genet. 2, 153–175 (2001).
Huang, T.N. et al. Tbr1 haploinsufficiency impairs amygdalar axonal projections and results in cognitive abnormality. Nat. Neurosci. 17, 240–247 (2014).
Chuang, H.C., Huang, T.N. & Hsueh, Y.P. Neuronal excitation upregulates Tbr1, a high-confidence risk gene of autism, mediating Grin2b expression in the adult brain. Front. Cell. Neurosci. 8, 280 (2014).
Flavell, S.W. et al. Activity-dependent regulation of MEF2 transcription factors suppresses excitatory synapse number. Science 311, 1008–1012 (2006).
Chen, W.G. et al. Derepression of BDNF transcription involves calcium-dependent phosphorylation of MeCP2. Science 302, 885–889 (2003).
Zhou, Z. et al. Brain-specific phosphorylation of MeCP2 regulates activity-dependent Bdnf transcription, dendritic growth and spine maturation. Neuron 52, 255–269 (2006).
Castro, J. et al. Functional recovery with recombinant human IGF1 treatment in a mouse model of Rett Syndrome. Proc. Natl. Acad. Sci. USA 111, 9941–9946 (2014).
Tropea, D. et al. Partial reversal of Rett syndrome–like symptoms in MeCP2-mutant mice. Proc. Natl. Acad. Sci. USA 106, 2029–2034 (2009).
Bozdagi, O., Tavassoli, T. & Buxbaum, J.D. Insulin-like growth factor 1 rescues synaptic and motor deficits in a mouse model of autism and developmental delay. Mol. Autism 4, 9 (2013).
Subramaniam, S. et al. Insulin-like growth factor 1 inhibits extracellular signal–regulated kinase to promote neuronal survival via the phosphatidylinositol 3-kinase–protein kinase A–c-Raf pathway. J. Neurosci. 25, 2838–2852 (2005).
Ehninger, D. & Silva, A.J. Rapamycin for treating tuberous sclerosis and autism spectrum disorders. Trends Mol. Med. 17, 78–87 (2011).
Bassell, G.J. & Warren, S.T. Fragile X syndrome: loss of local mRNA regulation alters synaptic development and function. Neuron 60, 201–214 (2008).
Napoli, I. et al. The fragile X syndrome protein represses activity-dependent translation through CYFIP1, a new 4E-BP. Cell 134, 1042–1054 (2008).
Nishimura, Y. et al. Genome-wide expression profiling of lymphoblastoid cell lines distinguishes different forms of autism and reveals shared pathways. Hum. Mol. Genet. 16, 1682–1698 (2007).
Tsai, P.T. et al. Autistic-like behavior and cerebellar dysfunction in Purkinje cell Tsc1-mutant mice. Nature 488, 647–651 (2012).
Zhou, J. et al. Pharmacological inhibition of mTORC1 suppresses anatomical, cellular and behavioral abnormalities in neural-specific Pten-knockout mice. J. Neurosci. 29, 1773–1783 (2009).
Gkogkas, C.G. et al. Autism-related deficits via dysregulated eIF4E-dependent translational control. Nature 493, 371–377 (2013).
Santini, E. et al. Exaggerated translation causes synaptic and behavioral aberrations associated with autism. Nature 493, 411–415 (2013).
Berg, J.M. et al. JAKMIP1, a novel regulator of neuronal translation, modulates synaptic function and autistic-like behaviors in mouse. Neuron 88, 1173–1191 (2015).
Darnell, J.C. et al. FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism. Cell 146, 247–261 (2011).
Huber, K.M., Gallagher, S.M., Warren, S.T. & Bear, M.F. Altered synaptic plasticity in a mouse model of fragile X mental retardation. Proc. Natl. Acad. Sci. USA 99, 7746–7750 (2002).
Cohen, S. & Greenberg, M.E. Communication between the synapse and the nucleus in neuronal development, plasticity and disease. Annu. Rev. Cell Dev. Biol. 24, 183–209 (2008).
Hua, J.Y. & Smith, S.J. Neural activity and the dynamics of central nervous system development. Nat. Neurosci. 7, 327–332 (2004).
Tsai, N.P. et al. Multiple autism-linked genes mediate synapse elimination via proteasomal degradation of a synaptic scaffold PSD-95. Cell 151, 1581–1594 (2012).
Wilkerson, J.R. et al. A role for dendritic mGluR5-mediated local translation of Arc (Arg 3.1) in MEF2-dependent synapse elimination. Cell Rep. 7, 1589–1600 (2014).
Abrahams, B.S. & Geschwind, D.H. Connecting genes to brain in the autism spectrum disorders. Arch. Neurol. 67, 395–399 (2010).
Yerys, B.E. & Herrington, J.D. Multimodal imaging in autism: an early review of comprehensive neural circuit characterization. Curr. Psychiatry Rep. 16, 496 (2014).
Luckhardt, C., Jarczok, T.A. & Bender, S. Elucidating the neurophysiological underpinnings of autism spectrum disorder: new developments. J. Neural Transm. 121, 1129–1144 (2014).
Ecker, C., Bookheimer, S.Y. & Murphy, D.G. Neuroimaging in autism spectrum disorder: brain structure and function across the lifespan. Lancet Neurol. 14, 1121–1134 (2015).
Minshew, N.J. & Williams, D.L. The new neurobiology of autism: cortex, connectivity and neuronal organization. Arch. Neurol. 64, 945–950 (2007).
Zikopoulos, B. & Barbas, H. Changes in prefrontal axons may disrupt the network in autism. J. Neurosci. 30, 14595–14609 (2010).
McPartland, J.C., Coffman, M. & Pelphrey, K.A. Recent advances in understanding the neural bases of autism spectrum disorder. Curr. Opin. Pediatr. 23, 628–632 (2011).
Rudie, J.D. et al. Autism-associated promoter variant in MET impacts functional and structural brain networks. Neuron 75, 904–915 (2012).
Amaral, D.G., Schumann, C.M. & Nordahl, C.W. Neuroanatomy of autism. Trends Neurosci. 31, 137–145 (2008).
Vissers, M.E., Cohen, M.X. & Geurts, H.M. Brain connectivity and high functioning autism: a promising path of research that needs refined models, methodological convergence and stronger behavioral links. Neurosci. Biobehav. Rev. 36, 604–625 (2012).
Ellegood, J. et al. Clustering autism: using neuroanatomical differences in 26 mouse models to gain insight into the heterogeneity. Mol. Psychiatry 20, 118–125 (2015).
Kloth, A.D. et al. Cerebellar associative sensory learning defects in five mouse autism models. eLife 4, e06085 (2015).
Wang, S.S., Kloth, A.D. & Badura, A. The cerebellum, sensitive periods and autism. Neuron 83, 518–532 (2014).
Amaral, D.G. The amygdala, social behavior and danger detection. Ann. NY Acad. Sci. 1000, 337–347 (2003).
Baron-Cohen, S. et al. The amygdala theory of autism. Neurosci. Biobehav. Rev. 24, 355–364 (2000).
Langen, M., Durston, S., Kas, M.J., van Engeland, H. & Staal, W.G. The neurobiology of repetitive behavior: ...and men. Neurosci. Biobehav. Rev. 35, 356–365 (2011).
Peça, J. et al. Shank3-mutant mice display autistic-like behaviors and striatal dysfunction. Nature 472, 437–442 (2011).
Rothwell, P.E. et al. Autism-associated neuroligin–3 mutations commonly impair striatal circuits to boost repetitive behaviors. Cell 158, 198–212 (2014).
Geschwind, D.H. & Rakic, P. Cortical evolution: judge the brain by its cover. Neuron 80, 633–647 (2013).
Kasthuri, N. et al. Saturated reconstruction of a volume of neocortex. Cell 162, 648–661 (2015).
Zingg, B. et al. Neural networks of the mouse neocortex. Cell 156, 1096–1111 (2014).
Gunaydin, L.A. et al. Natural neural projection dynamics underlying social behavior. Cell 157, 1535–1551 (2014).
Marlin, B.J., Mitre, M., D'amour, J.A., Chao, M.V. & Froemke, R.C. Oxytocin enables maternal behavior by balancing cortical inhibition. Nature 520, 499–504 (2015).
Morgan, J.T. et al. Microglial activation and increased microglial density observed in the dorsolateral prefrontal cortex in autism. Biol. Psychiatry 68, 368–376 (2010).
Suzuki, K. et al. Microglial activation in young adults with autism spectrum disorder. JAMA Psychiatry 70, 49–58 (2013).
Tetreault, N.A. et al. Microglia in the cerebral cortex in autism. J. Autism Dev. Disord. 42, 2569–2584 (2012).
Vargas, D.L., Nascimbene, C., Krishnan, C., Zimmerman, A.W. & Pardo, C.A. Neuroglial activation and neuroinflammation in the brain of patients with autism. Ann. Neurol. 57, 67–81 (2005).
Laurence, J.A. & Fatemi, S.H. Glial fibrillary acidic protein is elevated in superior frontal, parietal and cerebellar cortices of autistic subjects. Cerebellum 4, 206–210 (2005).
Lopez-Hurtado, E. & Prieto, J. A microscopic study of language-related cortex in autism. Am. J. Biochem. Biotechnol. 4, 130–145 (2008).
Gupta, S. et al. Transcriptome analysis reveals dysregulation of innate immune response genes and neuronal activity–dependent genes in autism. Nat. Commun. 5, 5748 (2014).
Paolicelli, R.C. et al. Synaptic pruning by microglia is necessary for normal brain development. Science 333, 1456–1458 (2011).
Zhan, Y. et al. Deficient neuron-microglia signaling results in impaired functional brain connectivity and social behavior. Nat. Neurosci. 17, 400–406 (2014).
Chung, W.S. et al. Astrocytes mediate synapse elimination through MEGF10 and MERTK pathways. Nature 504, 394–400 (2013).
Clarke, L.E. & Barres, B.A. Emerging roles of astrocytes in neural circuit development. Nat. Rev. Neurosci. 14, 311–321 (2013).
Schafer, D.P. et al. Microglia sculpt postnatal neural circuits in an activity- and complement-dependent manner. Neuron 74, 691–705 (2012).
de Vrij, F.M. et al. Rescue of behavioral phenotype and neuronal protrusion morphology in Fmr1 KO mice. Neurobiol. Dis. 31, 127–132 (2008).
Silverman, J.L., Tolu, S.S., Barkan, C.L. & Crawley, J.N. Repetitive self-grooming behavior in the BTBR mouse model of autism is blocked by the mGluR5 antagonist MPEP. Neuropsychopharmacology 35, 976–989 (2010).
Tian, D. et al. Contribution of mGluR5 to pathophysiology in a mouse model of human chromosome 16p11.2 microdeletion. Nat. Neurosci. 18, 182–184 (2015).
Won, H. et al. Autistic-like social behavior in Shank2-mutant mice improved by restoring NMDA receptor function. Nature 486, 261–265 (2012).
Yan, Q.J., Rammal, M., Tranfaglia, M. & Bauchwitz, R.P. Suppression of two major fragile X syndrome mouse model phenotypes by the mGluR5 antagonist MPEP. Neuropharmacology 49, 1053–1066 (2005).
Benson, A.D., Burket, J.A. & Deutsch, S.I. Balb/c mice treated with D-cycloserine arouse increased social interest in conspecifics. Brain Res. Bull. 99, 95–99 (2013).
Blundell, J. et al. Neuroligin-1 deletion results in impaired spatial memory and increased repetitive behavior. J. Neurosci. 30, 2115–2129 (2010).
Burket, J.A., Benson, A.D., Tang, A.H. & Deutsch, S.I. D-cycloserine improves sociability in the BTBR T+ Itpr3tf/J mouse model of autism spectrum disorders with altered Ras-Raf-ERK1/2 signaling. Brain Res. Bull. 96, 62–70 (2013).
Auerbach, B.D., Osterweil, E.K. & Bear, M.F. Mutations causing syndromic autism define an axis of synaptic pathophysiology. Nature 480, 63–68 (2011).
Han, S., Tai, C., Jones, C.J., Scheuer, T. & Catterall, W.A. Enhancement of inhibitory neurotransmission by GABAA receptors having a2,3-subunits ameliorates behavioral deficits in a mouse model of autism. Neuron 81, 1282–1289 (2014).
Han, S. et al. Autistic-like behaviour in Scn1a+/− mice and rescue by enhanced GABA-mediated neurotransmission. Nature 489, 385–390 (2012).
Ehninger, D. et al. Reversal of learning deficits in a Tsc2+/− mouse model of tuberous sclerosis. Nat. Med. 14, 843–848 (2008).
Oguro-Ando, A. et al. Increased CYFIP1 dosage alters cellular and dendritic morphology and dysregulates mTOR. Mol. Psychiatry 20, 1069–1078 (2015).
Ru, W., Peng, Y., Zhong, L. & Tang, S.J. A role of the mammalian target of rapamycin (mTOR) in glutamate-induced downregulation of tuberous sclerosis complex proteins 2 (TSC2). J. Mol. Neurosci. 47, 340–345 (2012).
Chang, Q., Khare, G., Dani, V., Nelson, S. & Jaenisch, R. The disease progression of Mecp2-mutant mice is affected by the level of BDNF expression. Neuron 49, 341–348 (2006).
Kline, D.D., Ogier, M., Kunze, D.L. & Katz, D.M. Exogenous brain-derived neurotrophic factor rescues synaptic dysfunction in Mecp2-null mice. J. Neurosci. 30, 5303–5310 (2010).
Mellios, N. et al. b2-adrenergic receptor agonist ameliorates phenotypes and corrects microRNA-mediated IGF1 deficits in a mouse model of Rett syndrome. Proc. Natl. Acad. Sci. USA 111, 9947–9952 (2014).
Deogracias, R. et al. Fingolimod, a sphingosine-1 phosphate receptor modulator, increases BDNF levels and improves symptoms of a mouse model of Rett syndrome. Proc. Natl. Acad. Sci. USA 109, 14230–14235 (2012).
Ross, H.E. & Young, L.J. Oxytocin and the neural mechanisms regulating social cognition and affiliative behavior. Front. Neuroendocrinol. 30, 534–547 (2009).
Insel, T.R. The challenge of translation in social neuroscience: a review of oxytocin, vasopressin and affiliative behavior. Neuron 65, 768–779 (2010).
Peñagarikano, O. et al. Exogenous and evoked oxytocin restores social behavior in the Cntnap2 mouse model of autism. Sci. Transl. Med. 7, 271ra8 (2015).
Sala, M. et al. Pharmacologic rescue of impaired cognitive flexibility, social deficits, increased aggression and seizure susceptibility in oxytocin receptor–null mice: a neurobehavioral model of autism. Biol. Psychiatry 69, 875–882 (2011).
Tyzio, R. et al. Oxytocin-mediated GABA inhibition during delivery attenuates autism pathogenesis in rodent offspring. Science 343, 675–679 (2014).
Owen, S.F. et al. Oxytocin enhances hippocampal spike transmission by modulating fast-spiking interneurons. Nature 500, 458–462 (2013).
Hsiao, E.Y. et al. Microbiota modulate behavioral and physiological abnormalities associated with neurodevelopmental disorders. Cell 155, 1451–1463 (2013).
Ey, E. et al. Absence of deficits in social behaviors and ultrasonic vocalizations in later generations of mice lacking neuroligin 4-like. Genes Brain Behav. 11, 928–941 (2012).
Bernardet, M. & Crusio, W.E. Fmr1 KO mice as a possible model of autistic features. ScientificWorldJournal 6, 1164–1176 (2006).
Chadman, K.K. et al. Minimal aberrant behavioral phenotypes of neuroligin-3R451C–knockin mice. Autism Res. 1, 147–158 (2008).
Jacquemont, S. et al. Epigenetic modification of the FMR1 gene in fragile X syndrome is associated with differential response to the mGluR5 antagonist AFQ056. Sci. Transl. Med. 3, 64ra1 (2011).
Tachibana, M. et al. Long-term administration of intranasal oxytocin is a safe and promising therapy for early adolescent boys with autism spectrum disorders. J. Child Adolesc. Psychopharmacol. 23, 123–127 (2013).
Dadds, M.R. et al. Nasal oxytocin for social deficits in childhood autism: a randomized controlled trial. J. Autism Dev. Disord. 44, 521–531 (2014).
Guastella, A.J. et al. The effects of a course of intranasal oxytocin on social behaviors in youth diagnosed with autism spectrum disorders: a randomized controlled trial. J. Child Psychol. Psychiatry 56, 444–452 (2015).
Jeste, S.S. & Geschwind, D.H. Clinical trials for neurodevelopmental disorders: at a therapeutic frontier. Sci. Transl. Med. 8, 321fs1 (2016).
Bailey, K.R., Rustay, N.R. & Crawley, J.N. Behavioral phenotyping of transgenic and knockout mice: practical concerns and potential pitfalls. ILAR J. 47, 124–131 (2006).
Werling, D.M., Lowe, J.K., Luo, R., Cantor, R.M. & Geschwind, D.H. Replication of linkage at chromosome 20p13 and identification of suggestive sex-differential risk loci for autism spectrum disorder. Mol. Autism 5, 13 (2014).
Cloughesy, T.F., Cavenee, W.K. & Mischel, P.S. Glioblastoma: from molecular pathology to targeted treatment. Annu. Rev. Pathol. 9, 1–25 (2014).
Werker, J.F. & Hensch, T.K. Critical periods in speech perception: new directions. Annu. Rev. Psychol. 66, 173–196 (2015).
Falconer, D.S. The inheritance of liability to certain diseases, estimated from the incidence among relatives. Ann. Hum. Genet. 29, 51–76 (1965).
Isshiki, M. et al. Enhanced synapse remodeling as a common phenotype in mouse models of autism. Nat. Commun. 5, 4742 (2014).
Nakatani, J. et al. Abnormal behavior in a chromosome-engineered mouse model for human 15q11–13 duplication seen in autism. Cell 137, 1235–1246 (2009).
Lee, E.J. et al. Trans-synaptic zinc mobilization improves social interaction in two mouse models of autism through NMDAR activation. Nat. Commun. 6, 7168 (2015).
Lim, C.S. et al. Pharmacological rescue of Ras signaling, GluA1-dependent synaptic plasticity and learning deficits in a fragile X model. Genes Dev. 28, 273–289 (2014).
Gross, C. et al. Selective role of the catalytic PI3K subunit p110-β in impaired higher-order cognition in fragile X syndrome. Cell Reports 11, 681–688 (2015).
Osterweil, E.K., Krueger, D.D., Reinhold, K. & Bear, M.F. Hypersensitivity to mGluR5 and ERK1/2 leads to excessive protein synthesis in the hippocampus of a mouse model of fragile X syndrome. J. Neurosci. 30, 15616–15627 (2010).
Etherton, M.R., Blaiss, C.A., Powell, C.M. & Südhof, T.C. Mouse neurexin I alpha deletion causes correlated electrophysiological and behavioral changes consistent with cognitive impairments. Proc. Natl. Acad. Sci. USA 106, 17998–18003 (2009).
Grayton, H.M., Missler, M., Collier, D.A. & Fernandes, C. Altered social behaviors in neurexin-1α–knockout mice resemble core symptoms in neurodevelopmental disorders. PLoS One 8, e67114 (2013).
Tabuchi, K. et al. A neuroligin-3 mutation implicated in autism increases inhibitory synaptic transmission in mice. Science 318, 71–76 (2007).
Etherton, M. et al. Autism-linked neuroligin-3R451C mutation differentially alters hippocampal and cortical synaptic function. Proc. Natl. Acad. Sci. USA 108, 13764–13769 (2011).
Baudouin, S.J. et al. Shared synaptic pathophysiology in syndromic and nonsyndromic rodent models of autism. Science 338, 128–132 (2012).
Jamain, S. et al. Reduced social interaction and ultrasonic communication in a mouse model of monogenic heritable autism. Proc. Natl. Acad. Sci. USA 105, 1710–1715 (2008).
Schmeisser, M.J. et al. Autistic-like behaviors and hyperactivity in mice lacking ProSAP1 (Shank2). Nature 486, 256–260 (2012).
Wang, X. et al. Synaptic dysfunction and abnormal behaviors in mice lacking major isoforms of Shank3. Hum. Mol. Genet. 20, 3093–3108 (2011).
Bozdagi, O. et al. Haploinsufficiency of the autism-associated Shank3 gene leads to deficits in synaptic function, social interaction and social communication. Mol. Autism 1, 15 (2010).
Duffney, L.J. et al. Autism-like deficits in Shank3-deficient mice are rescued by targeting actin regulators. Cell Rep. 11, 1400–1413 (2015).
Gdalyahu, A. et al. The autism-related protein contactin-associated protein-like 2 (CNTNAP2) stabilizes new spines: an in vivo mouse study. PLoS One 10, e0125633 (2015).
Derecki, N.C. et al. Wild-type microglia arrest pathology in a mouse model of Rett syndrome. Nature 484, 105–109 (2012).
Lioy, D.T. et al. A role for glia in the progression of Rett's syndrome. Nature 475, 497–500 (2011).
Langen, M., Kas, M.J., Staal, W.G., van Engeland, H. & Durston, S. The neurobiology of repetitive behavior: of mice.... Neurosci. Biobehav. Rev. 35, 345–355 (2011).
Lui, J.H. et al. Radial glia require PDGFD–PDGFR-β signaling in human but not mouse neocortex. Nature 515, 264–268 (2014).
Merkle, F.T. & Eggan, K. Modeling human disease with pluripotent stem cells: from genome association to function. Cell Stem Cell 12, 656–668 (2013).
Griesi-Oliveira, K. et al. Modeling nonsyndromic autism and the impact of TRPC6 disruption in human neurons. Mol. Psychiatry 20, 1350–1365 (2015).
Ricciardi, S. et al. CDKL5 ensures excitatory synapse stability by reinforcing NGL-1–PSD95 interaction in the postsynaptic compartment and is impaired in patient iPSC-derived neurons. Nat. Cell Biol. 14, 911–923 (2012).
Kadoshima, T. et al. Self-organization of axial polarity, inside-out layer pattern and species-specific progenitor dynamics in human ES cell–derived neocortex. Proc. Natl. Acad. Sci. USA 110, 20284–20289 (2013).
Lepski, G. et al. Delayed functional maturation of human neuronal progenitor cells in vitro. Mol. Cell. Neurosci. 47, 36–44 (2011).
Mariani, J. et al. Modeling human cortical development in vitro using induced pluripotent stem cells. Proc. Natl. Acad. Sci. USA 109, 12770–12775 (2012).
Nicholas, C.R. et al. Functional maturation of hPSC-derived forebrain interneurons requires an extended timeline and mimics human neural development. Cell Stem Cell 12, 573–586 (2013).
van de Leemput, J. et al. CORTECON: a temporal transcriptome analysis of in vitro human cerebral cortex development from human embryonic stem cells. Neuron 83, 51–68 (2014).
Ben-David, E. & Shifman, S. Combined analysis of exome sequencing points toward a major role for transcription regulation during brain development in autism. Mol. Psychiatry 18, 1054–1056 (2013).
Paşca, A.M. et al. Functional cortical neurons and astrocytes from human pluripotent stem cells in 3D culture. Nat. Methods 12, 671–678 (2015).
Espuny-Camacho, I. et al. Pyramidal neurons derived from human pluripotent stem cells integrate efficiently into mouse brain circuits in vivo. Neuron 77, 440–456 (2013).
Hansen, D.V., Lui, J.H., Parker, P.R. & Kriegstein, A.R. Neurogenic radial glia in the outer subventricular zone of human neocortex. Nature 464, 554–561 (2010).
Lancaster, M.A. et al. Cerebral organoids model human brain development and microcephaly. Nature 501, 373–379 (2013).
Acknowledgements
We thank members of the Geschwind laboratory for helpful discussions and critical reading of the manuscript. This work was supported by US National Institutes of Health (NIH) grants 5R37 MH060233 (D.H.G.), 5R01 MH094714 (D.H.G.) and K99MH102357 (J.L.S.), the California Institute for Regenerative Medicine (CIRM)–Broad Stem Cell Research Center (BSCRC) training grant TG2-01169 (L.d.l.T.-U.) and the Glenn–American Federation for Aging Research (AFAR) Postdoctoral Fellowship Program for Translational Research on Aging award 20145357 (H.W.).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Table 1
Genetic contributions to autism. (XLSX 18 kb)
Supplementary Table 2
Mouse models of ASD. (XLSX 42 kb)
Supplementary Table 3
Human in vitro models of ASD. (XLSX 42 kb)
Rights and permissions
About this article
Cite this article
de la Torre-Ubieta, L., Won, H., Stein, J. et al. Advancing the understanding of autism disease mechanisms through genetics. Nat Med 22, 345–361 (2016). https://doi.org/10.1038/nm.4071
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nm.4071
This article is cited by
-
Remodeling of the postsynaptic proteome in male mice and marmosets during synapse development
Nature Communications (2024)
-
Possible roles of deep cortical neurons and oligodendrocytes in the neural basis of human sociality
Anatomical Science International (2024)
-
Neurophysiological measures of auditory sensory processing are associated with adaptive behavior in children with Autism Spectrum Disorder
Journal of Neurodevelopmental Disorders (2023)
-
Tsc2 mutation rather than Tsc1 mutation dominantly causes a social deficit in a mouse model of tuberous sclerosis complex
Human Genomics (2023)
-
Adaptive control of synaptic plasticity integrates micro- and macroscopic network function
Neuropsychopharmacology (2023)