Abstract
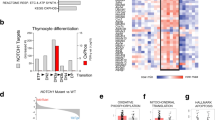
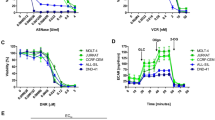
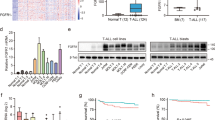
Activating mutations in NOTCH1 are common in T cell acute lymphoblastic leukemia (T-ALL). Here we identify glutaminolysis as a critical pathway for leukemia cell growth downstream of NOTCH1 and a key determinant of the response to anti-NOTCH1 therapies in vivo. Mechanistically, inhibition of NOTCH1 signaling in T-ALL induces a metabolic shutdown, with prominent inhibition of glutaminolysis and triggers autophagy as a salvage pathway supporting leukemia cell metabolism. Consequently, inhibition of glutaminolysis and inhibition of autophagy strongly and synergistically enhance the antileukemic effects of anti-NOTCH1 therapy in mice harboring T-ALL. Moreover, we demonstrate that Pten loss upregulates glycolysis and consequently rescues leukemic cell metabolism, thereby abrogating the antileukemic effects of NOTCH1 inhibition. Overall, these results identify glutaminolysis as a major node in cancer metabolism controlled by NOTCH1 and as therapeutic target for the treatment of T-ALL.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Change history
23 October 2015
Mouse images are duplicated in Fig. 6e (day 0 Vehicle and day 0 BPTES) and in Fig. 6f (day 0 DBZ + BPTES and day 6 DBZ + BPTES). The authors made these errors in assembling the figure panels. The authors have now supplied corrected versions of these panels, in which the correct micrographs for Fig. 6e (day 0 BPTES) and Fig. 6f (day 6 DBZ + BPTS) are included. These errors do not affect the data shown in the graphs in Fig. 6e,f. The errors have been corrected in the HTML and PDF versions of the article.
References
Hori, K., Sen, A. & Artavanis-Tsakonas, S. Notch signaling at a glance. J. Cell Sci. 126, 2135–2140 (2013).
Rothenberg, E.V., Moore, J.E. & Yui, M.A. Launching the T-cell-lineage developmental programme. Nat. Rev. Immunol. 8, 9–21 (2008).
Thompson, P.K. & Zuniga-Pflucker, J.C. On becoming a T cell, a convergence of factors kick it up a Notch along the way. Semin. Immunol. 23, 350–359 (2011).
Ciofani, M. & Zuniga-Pflucker, J.C. Notch promotes survival of pre-T cells at the beta-selection checkpoint by regulating cellular metabolism. Nat. Immunol. 6, 881–888 (2005).
Weng, A.P. et al. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science 306, 269–271 (2004).
Selkoe, D. & Kopan, R. Notch and presenilin: regulated intramembrane proteolysis links development and degeneration. Annu. Rev. Neurosci. 26, 565–597 (2003).
Palomero, T. et al. Mutational loss of PTEN induces resistance to NOTCH1 inhibition in T-cell leukemia. Nat. Med. 13, 1203–1210 (2007).
Chiang, M.Y. et al. Leukemia-associated NOTCH1 alleles are weak tumor initiators but accelerate K-ras-initiated leukemia. J. Clin. Invest. 118, 3181–3194 (2008).
Milano, J. et al. Modulation of notch processing by γ-secretase inhibitors causes intestinal goblet cell metaplasia and induction of genes known to specify gut secretory lineage differentiation. Toxicol. Sci. 82, 341–358 (2004).
Palomero, T. et al. NOTCH1 directly regulates c-MYC and activates a feed-forward-loop transcriptional network promoting leukemic cell growth. Proc. Natl. Acad. Sci. USA 103, 18261–18266 (2006).
Buzzai, M. et al. The glucose dependence of Akt-transformed cells can be reversed by pharmacologic activation of fatty acid β-oxidation. Oncogene 24, 4165–4173 (2005).
Wise, D.R. et al. Myc regulates a transcriptional program that stimulates mitochondrial glutaminolysis and leads to glutamine addiction. Proc. Natl. Acad. Sci. USA 105, 18782–18787 (2008).
Komatsu, M. et al. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J. Cell Biol. 169, 425–434 (2005).
Schroeter, E.H., Kisslinger, J.A. & Kopan, R. Notch-1 signalling requires ligand-induced proteolytic release of intracellular domain. Nature 393, 382–386 (1998).
Locasale, J.W. et al. Phosphoglycerate dehydrogenase diverts glycolytic flux and contributes to oncogenesis. Nat. Genet. 43, 869–874 (2011).
Robinson, M.M. et al. Novel mechanism of inhibition of rat kidney-type glutaminase by bis-2-(5-phenylacetamido-1,2,4-thiadiazol-2-yl)ethyl sulfide (BPTES). Biochem. J. 406, 407–414 (2007).
Tennant, D.A., Duran, R.V. & Gottlieb, E. Targeting metabolic transformation for cancer therapy. Nat. Rev. Cancer 10, 267–277 (2010).
Homminga, I. et al. Characterization of a pediatric T-cell acute lymphoblastic leukemia patient with simultaneous LYL1 and LMO2 rearrangements. Haematologica 97, 258–261 (2012).
Medyouf, H. et al. Acute T-cell leukemias remain dependent on Notch signaling despite PTEN and INK4A/ARF loss. Blood 115, 1175–1184 (2010).
Knoechel, B. et al. An epigenetic mechanism of resistance to targeted therapy in T cell acute lymphoblastic leukemia. Nat. Genet. 46, 364–370 (2014).
Ying, H. et al. Oncogenic Kras maintains pancreatic tumors through regulation of anabolic glucose metabolism. Cell 149, 656–670 (2012).
Zhou, S. et al. Autophagy in tumorigenesis and cancer therapy: Dr. Jekyll or Mr. Hyde? Cancer Lett. 323, 115–127 (2012).
Elstrom, R.L. et al. Akt stimulates aerobic glycolysis in cancer cells. Cancer Res. 64, 3892–3899 (2004).
Bauer, D.E., Hatzivassiliou, G., Zhao, F., Andreadis, C. & Thompson, C.B. ATP citrate lyase is an important component of cell growth and transformation. Oncogene 24, 6314–6322 (2005).
Hatzivassiliou, G. et al. ATP citrate lyase inhibition can suppress tumor cell growth. Cancer Cell 8, 311–321 (2005).
Tong, X., Zhao, F. & Thompson, C.B. The molecular determinants of de novo nucleotide biosynthesis in cancer cells. Curr. Opin. Genet. Dev. 19, 32–37 (2009).
Guo, K. et al. Disruption of peripheral leptin signaling in mice results in hyperleptinemia without associated metabolic abnormalities. Endocrinology 148, 3987–3997 (2007).
Trotman, L.C. et al. Pten dose dictates cancer progression in the prostate. PLoS Biol. 1, e59 (2003).
Kimbrel, E.A., Davis, T.N., Bradner, J.E. & Kung, A.L. In vivo pharmacodynamic imaging of proteasome inhibition. Mol. Imaging 8, 140–147 (2009).
Gentleman, R.C. et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 5, R80 (2004).
Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 102, 15545–15550 (2005).
Campos-Sandoval, J.A. et al. Expression of functional human glutaminase in baculovirus system: affinity purification, kinetic and molecular characterization. Int. J. Biochem. Cell Biol. 39, 765–773 (2007).
Heckl, D. et al. Generation of mouse models of myeloid malignancy with combinatorial genetic lesions using CRISPR-Cas9 genome editing. Nat. Biotechnol. 32, 941–946 (2014).
Chou, T.C. Drug combination studies and their synergy quantification using the Chou-Talalay method. Cancer Res. 70, 440–446 (2010).
Acknowledgements
We are grateful to J. Aster (Brigham and Women's Hospital, Harvard Medical School) for the MigR1-NOTCH1 L1601P ΔPEST vector, D. Vignali (University of Pittsburgh) for the pMSCV-mCherry FP vector, W. Pear (Abramson Family Cancer Research Institute, University of Pennsylvania) for the MigR1 vector, B. Ebert (Dana-Farber Cancer Research Institute) for the pL-CRISPR.EFS.GFP vector, P. Pandolfi (Beth Israel Deaconess Medical Center, Harvard Medical School) for the Pten conditional knockout mouse, T. Ludwig (Columbia University Medical Center) for the Rosa26Cre-ERT2/+ mouse, M. Komatsu (Tokyo Metropolitan Institute of Medical Science) for the Atg7 conditional knockout mouse, S. Indraccolo (Istituto di Ricovero e Cura a Carattere Scientifico) for xenograft T-ALL cells and R. Baer and C. Lopez-Otin for helpful discussions and revision of the manuscript. This work was supported by the US National Institutes of Health grants R01CA129382 and CA120196, the Stand Up To Cancer Innovative Research Award and the Swim Across America Foundation (A.A.F.). J.M. and J.M.M. were supported by CVI-6656 (Junta de Andalucía, Spain). D.H. is supported by the Leukemia and Lymphoma Society. M.S.-M. and A.A.W. are supported by the Rally Foundation. L.B. is supported by the Lymphoma Research Foundation.
Author information
Authors and Affiliations
Contributions
D.H. carried out most of the experiments. A.A.-I. analyzed gene expression profile signatures. J.S. performed metabolic studies. M.S.-M. performed some in vivo and in vitro drug response analyses. L.B. analyzed PTEN levels by intracellular FACS staining. V.T. generated some of the NOTCH1-induced primary leukemias. L.X. performed some animal studies with D.H. A.A.W. performed some experiments with human primary T-ALL samples. M.C. conducted histological evaluation of tumor development and response to therapy. J.E.H. performed some in vivo experiments. J.M. and J.M.M. contributed reagents. S.R. generated the Gls conditional knockout mice. A.L.K. conceived and supervised bioimaging studies. C.C.-C. supervised histological analyses. R.J.D. supervised metabolic isotope tracing analyses. A.A.F designed the study, supervised research and wrote the manuscript with D.H.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–14 and Supplementary Table 1 (PDF 2395 kb)
Rights and permissions
About this article
Cite this article
Herranz, D., Ambesi-Impiombato, A., Sudderth, J. et al. Metabolic reprogramming induces resistance to anti-NOTCH1 therapies in T cell acute lymphoblastic leukemia. Nat Med 21, 1182–1189 (2015). https://doi.org/10.1038/nm.3955
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nm.3955
This article is cited by
-
Slow TCA flux and ATP production in primary solid tumours but not metastases
Nature (2023)
-
Valine tRNA levels and availability regulate complex I assembly in leukaemia
Nature (2022)
-
Notch signaling pathway: architecture, disease, and therapeutics
Signal Transduction and Targeted Therapy (2022)
-
Inhibition of mitochondrial complex I reverses NOTCH1-driven metabolic reprogramming in T-cell acute lymphoblastic leukemia
Nature Communications (2022)
-
Notch3 inhibits cell proliferation and tumorigenesis and predicts better prognosis in breast cancer through transactivating PTEN
Cell Death & Disease (2021)