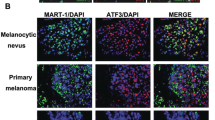
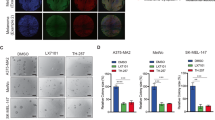
Abstract
Metastasis is responsible for most cancer-related deaths, and, among common tumor types, melanoma is one with great potential to metastasize. Here we study the contribution of epigenetic changes to the dissemination process by analyzing the changes that occur at the DNA methylation level between primary cancer cells and metastases. We found a hypomethylation event that reactivates a cryptic transcript of the Rab GTPase activating protein TBC1D16 (TBC1D16-47 kDa; referred to hereafter as TBC1D16-47KD) to be a characteristic feature of the metastatic cascade. This short isoform of TBC1D16 exacerbates melanoma growth and metastasis both in vitro and in vivo. By combining immunoprecipitation and mass spectrometry, we identified RAB5C as a new TBC1D16 target and showed that it regulates EGFR in melanoma cells. We also found that epigenetic reactivation of TBC1D16-47KD is associated with poor clinical outcome in melanoma, while conferring greater sensitivity to BRAF and MEK inhibitors.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Accession codes
References
Siegel, R., Naishadham, D. & Jemal, A. Cancer statistics. CA Cancer J. Clin. 63, 11–30 (2013).
Jones, P.A. & Baylin, S.B. The epigenomics of cancer. Cell 128, 683–692 (2007).
Heyn, H. & Esteller, M. DNA methylation profiling in the clinic: applications and challenges. Nat. Rev. Genet. 13, 679–692 (2012).
Timp, W. & Feinberg, A.P. Cancer as a dysregulated epigenome allowing cellular growth advantage at the expense of the host. Nat. Rev. Cancer 13, 497–510 (2013).
Fang, F. et al. Breast cancer methylomes establish an epigenomic foundation for metastasis. Sci. Transl. Med. 3, 75ra25 (2011).
Cunha, S. et al. The RON receptor tyrosine kinase promotes metastasis by triggering MBD4-dependent DNA methylation reprogramming. Cell Reports 6, 141–154 (2014).
Carmona, F.J. et al. A comprehensive DNA methylation profile of epithelial-to-mesenchymal transition. Cancer Res. 74, 5608–5619 (2014).
Lujambio, A. et al. A microRNA DNA methylation signature for human cancer metastasis. Proc. Natl. Acad. Sci. USA 105, 13556–13561 (2008).
Harbst, K. et al. Multiple metastases from cutaneous malignant melanoma patients may display heterogeneous genomic and epigenomic patterns. Melanoma Res. 20, 381–391 (2010).
Marzese, D.M. et al. Epigenome-wide DNA methylation landscape of melanoma progression to brain metastasis reveals aberrations on homeobox D cluster associated with prognosis. Hum. Mol. Genet. 23, 226–238 (2014).
MacKie, R.M., Hauschild, A. & Eggermont, A.M. Epidemiology of invasive cutaneous melanoma. Ann. Oncol. 20 (suppl. 6), vi1–vi7 (2009).
Villanueva, M.T. Skin cancer: in melanoma ulceration, size matters. Nat. Rev. Clin. Oncol 9, 370 (2012).
Mandalà, M. & Massi, D. Tissue prognostic biomarkers in primary cutaneous melanoma. Virchows Arch. 464, 265–281 (2014).
Tsao, H., Chin, L., Garraway, L.A. & Fisher, D.E. Melanoma: from mutations to medicine. Genes Dev. 26, 1131–1155 (2012).
Kaufman, H.L. et al. The Society for Immunotherapy of Cancer consensus statement on tumour immunotherapy for the treatment of cutaneous melanoma. Nat. Rev. Clin. Oncol 10, 588–598 (2013).
Kaufman, H.L. Melanoma as a model for precision medicine in oncology. Lancet Oncol. 15, 251–253 (2014).
Weinreb, A. & Travo, P. Discrimination between human melanoma cell lines by fluorescence anisotropy. Eur. J. Cancer Clin. Oncol. 20, 673–677 (1984).
Sandoval, J. et al. Validation of a DNA methylation microarray for 450,000 CpG sites in the human genome. Epigenetics 6, 692–702 (2011).
Akavia, U.D. et al. An integrated approach to uncover drivers of cancer. Cell 143, 1005–1017 (2010).
Goueli, B.S., Powell, M.B., Finger, E.C. & Pfeffer, S.R. TBC1D16 is a Rab4A GTPase activating protein that regulates receptor recycling and EGF receptor signaling. Proc. Natl. Acad. Sci. USA 109, 15787–15792 (2012).
El Kasmi, K.C. et al. Cutting edge: a transcriptional repressor and corepressor induced by the STAT3-regulated anti-inflammatory signalling pathway. J. Immunol. 179, 7215–7219 (2007).
Eisenberg, M.C. et al. Mechanistic modeling of the effects of myoferlin on tumor cell invasion. Proc. Natl. Acad. Sci. USA 108, 20078–20083 (2011).
Frasa, M.A., Koessmeier, K.T., Ahmadian, M.R. & Braga, V.M. Illuminating the functional and structural repertoire of human TBC/RABGAPs. Nat. Rev. Mol. Cell Biol. 13, 67–73 (2012).
Onodera, Y. et al. Rab5c promotes AMAP1–PRKD2 complex formation to enhance β1 integrin recycling in EGF-induced cancer invasion. J. Cell Biol. 197, 983–996 (2012).
Corcoran, R.B. et al. EGFR-mediated re-activation of MAPK signaling contributes to insensitivity of BRAF mutant colorectal cancers to RAF inhibition with vemurafenib. Cancer Discov. 2, 227–235 (2012).
Prahallad, A. et al. Unresponsiveness of colon cancer to BRAF(V600E) inhibition through feedback activation of EGFR. Nature 483, 100–103 (2012).
Yoon, Y.K. et al. Combination of EGFR and MEK1/2 inhibitor shows synergistic effects by suppressing EGFR/HER3-dependent AKT activation in human gastric cancer cells. Mol. Cancer Ther. 8, 2526–2536 (2009).
Mirzoeva, O.K. et al. Basal subtype and MAPK/ERK kinase (MEK)-phosphoinositide 3-kinase feedback signaling determine susceptibility of breast cancer cells to MEK inhibition. Cancer Res. 69, 565–572 (2009).
Garnett, M.J. et al. Systematic identification of genomic markers of drug sensitivity in cancer cells. Nature 483, 570–575 (2012).
Hansen, K.D. et al. Increased methylation variation in epigenetic domains across cancer types. Nat. Genet. 43, 768–775 (2011).
Hon, G.C. et al. Global DNA hypomethylation coupled to repressive chromatin domain formation and gene silencing in breast cancer. Genome Res. 22, 246–258 (2012).
Bert, S.A. et al. Regional activation of the cancer genome by long-range epigenetic remodeling. Cancer Cell 23, 9–22 (2013).
Kedlaya, R. et al. Interactions between GIPC-APPL and GIPC-TRP1 regulate melanosomal protein trafficking and melanogenesis in human melanocytes. Arch. Biochem. Biophys. 508, 227–233 (2011).
Huang, Z.M. et al. Targeting protein-trafficking pathways alters melanoma treatment sensitivity. Proc. Natl. Acad. Sci. USA 109, 553–558 (2012).
Gray-Schopfer, V., Wellbrock, C. & Marais, R. Melanoma biology and new targeted therapy. Nature 445, 851–857 (2007).
Middleton, M.R. et al. Randomized phase III study of temozolomide versus dacarbazine in the treatment of patients with advanced metastatic malignant melanoma. J. Clin. Oncol. 18, 158–166 (2000).
Eggermont, A.M., Spatz, A. & Robert, C. Cutaneous melanoma. Lancet 383, 816–827 (2014).
Catalanotti, F. et al. Phase II trial of MEK inhibitor selumetinib (AZD6244, ARRY-142886) in patients with BRAFV600E/K-mutated melanoma. Clin. Cancer Res. 19, 2257–2264 (2013).
Girotti, M.R. et al. Inhibiting EGF receptor or SRC family kinase signaling overcomes BRAF inhibitor resistance in melanoma. Cancer Discov. 3, 158–167 (2013).
Altman, D.G. et al. Reporting Recommendations for Tumor Marker Prognostic Studies (REMARK): explanation and elaboration. PLoS Med. 9, e1001216 (2012).
Heyn, H. et al. Distinct DNA methylomes of newborns and centenarians. Proc. Natl. Acad. Sci. USA 109, 10522–10527 (2012).
Moutinho, C. et al. Epigenetic inactivation of the BRCA1 interactor SRBC and resistance to oxaliplatin in colorectal cancer. J. Natl. Cancer Inst. 106, djt322 (2014).
Lopez-Serra, P. et al. A DERL3-associated defect in the degradation of SLC2A1 mediates the Warburg effect. Nat. Commun. 5, 3608 (2014).
Howard-Jones, N. A CIOMS ethical code for animal experimentation. WHO Chron. 39, 51–56 (1985).
Acknowledgements
We thank the patients and their families. The research leading to these results has received funding from the European Community's Seventh Framework Programme FP7/2007-2013 under grant agreement no. PIAPP-GA-2009-230614–Target-Melanoma project (F.P., J.O., W.M.G., M.E.), the Worldwide Cancer Research grant reference no. 15-0354 (M.E.), the European Research Council Advanced grant no. 268626–EPINORC project (M.E.), the Ministerio de Ciencia e Innovacion grant numbers SAF2011-22803 (M.E.) and FIS PI13-01339 (A. Villanueva), the CRUK Manchester Institute (C5759/A12328 to R.M.), the Wellcome Trust (100282/Z/12/Z to R.M.), the Cellex Foundation (M.E.) and the Health and Science Departments of the Catalan Government Generalitat de Catalunya 2005-SGR00727 (A. Villanueva) and 2014-SGR 633 (M.E.). M.V. was supported by a Formacion de Profesorado Universitario fellowship from the Spanish Ministry of Education. We thank the staff of the Animal Core Facility of Bellvitge Biomedical Research Institute for mouse care and maintenance. M.E. is an Institucio Catalana de Recerca i Estudis Avançats Research Professor.
Author information
Authors and Affiliations
Contributions
M.V. and M.E. conceived the study and wrote the manuscript. M.V. performed most experiments with the help of H.J.F., P.L.-S., F.J.C., S.G., C.M., J.L., A.P., H.H. and S.M. A.Vidal and A. Villanueva, together with M.M.-I., performed the mouse studies. A.M.-C., M.R.G., J.L.M., M.T.F.-F., E.E., E.M.-C., R.B.-E., A.B., F.P., J.v.d.O., W.M.G., D.T.F., K.T.F., U.M., P.L. and R.M. analyzed the clinical outcome and drug response data and provided conceptual input. All authors discussed the results and commented on the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Figures and Text
Supplementary Figures 1–12 & Supplementary Tables 1–3 (PDF 3464 kb)
Supplementary Data 1
2,620 CpGs most divergent between primary and metatatic tumor cell lines. (XLSX 322 kb)
Supplementary Data 2
CpGs located outside CpG islands most divergent between primary and metastatic tumor cell lines. (XLSX 12 kb)
Supplementary Data 3
DNA methylation profile of the TBC1D16-45/47KD promoter CpG island according to the DNA methylation microarray values in 36 melanoma cell lines. (XLSX 14 kb)
Rights and permissions
About this article
Cite this article
Vizoso, M., Ferreira, H., Lopez-Serra, P. et al. Epigenetic activation of a cryptic TBC1D16 transcript enhances melanoma progression by targeting EGFR. Nat Med 21, 741–750 (2015). https://doi.org/10.1038/nm.3863
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nm.3863
This article is cited by
-
Patient-specific identification of genome-wide DNA-methylation differences between intracranial and extracranial melanoma metastases
Scientific Reports (2023)
-
An epigenetic switch controls an alternative NR2F2 isoform that unleashes a metastatic program in melanoma
Nature Communications (2023)
-
Epigenetically regulated PCDHB15 impairs aggressiveness of metastatic melanoma cells
Clinical Epigenetics (2022)
-
Construction of mRNA prognosis signature associated with differentially expressed genes in early stage of stomach adenocarcinomas based on TCGA and GEO datasets
European Journal of Medical Research (2022)
-
Melanopsin (Opn4) is an oncogene in cutaneous melanoma
Communications Biology (2022)