Abstract
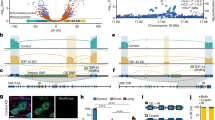
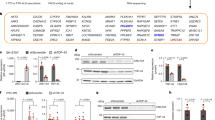
We have identified a rare coding mutation, T835M (rs137875858), in the UNC5C netrin receptor gene that segregated with disease in an autosomal dominant pattern in two families enriched for late-onset Alzheimer's disease and that was associated with disease across four large case-control cohorts (odds ratio = 2.15, Pmeta = 0.0095). T835M alters a conserved residue in the hinge region of UNC5C, and in vitro studies demonstrate that this mutation leads to increased cell death in human HEK293T cells and in rodent neurons. Furthermore, neurons expressing T835M UNC5C are more susceptible to cell death from multiple neurotoxic stimuli, including β-amyloid (Aβ), glutamate and staurosporine. On the basis of these data and the enriched hippocampal expression of UNC5C in the adult nervous system, we propose that one possible mechanism in which T835M UNC5C contributes to the risk of Alzheimer's disease is by increasing susceptibility to neuronal cell death, particularly in vulnerable regions of the Alzheimer's disease brain.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Coppola, G. et al. Evidence for a role of the rare p.A152T variant in MAPT in increasing the risk for FTD-spectrum and Alzheimer's diseases. Hum. Mol. Genet. 21, 3500–3512 (2012).
Corder, E.H. et al. Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer's disease in late onset families. Science 261, 921–923 (1993).
Cruchaga, C. et al. Rare variants in APP, PSEN1 and PSEN2 increase risk for AD in late-onset Alzheimer's disease families. PLoS ONE 7, e31039 (2012).
Cruchaga, C. et al. Rare coding variants in the phospholipase D3 gene confer risk for Alzheimer's disease. Nature 505, 550–554 (2014).
Genin, E. et al. APOE and Alzheimer disease: a major gene with semi-dominant inheritance. Mol. Psychiatry 16, 903–907 (2011).
Guerreiro, R. et al. TREM2 variants in Alzheimer's disease. N. Engl. J. Med. 368, 117–127 (2013).
Harold, D. et al. Genome-wide association study identifies variants at CLU and PICALM associated with Alzheimer's disease. Nat. Genet. 41, 1088–1093 (2009).
Hollingworth, P. et al. Common variants at ABCA7, MS4A6A/MS4A4E, EPHA1, CD33 and CD2AP are associated with Alzheimer's disease. Nat. Genet. 43, 429–435 (2011).
Jonsson, T. et al. A mutation in APP protects against Alzheimer's disease and age-related cognitive decline. Nature 488, 96–99 (2012).
Jonsson, T. et al. Variant of TREM2 associated with the risk of Alzheimer's disease. N. Engl. J. Med. 368, 107–116 (2013).
Lambert, J.C. et al. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer's disease. Nat. Genet. 45, 1452–1458 (2013).
Naj, A.C. et al. Common variants at MS4A4/MS4A6E, CD2AP, CD33 and EPHA1 are associated with late-onset Alzheimer's disease. Nat. Genet. 43, 436–441 (2011).
Harms, M. et al. C9orf72 hexanucleotide repeat expansions in clinical Alzheimer disease. JAMA Neurol. 70, 736–741 (2013).
Leonardo, E.D. et al. Vertebrate homologues of C. elegans UNC-5 are candidate netrin receptors. Nature 386, 833–838 (1997).
Ackerman, S.L. et al. The mouse rostral cerebellar malformation gene encodes an UNC-5-like protein. Nature 386, 838–842 (1997).
Przyborski, S.A., Knowles, B. & Ackerman, S. Embryonic phenotype of Unc5h3 mutant mice suggests chemorepulsion during the formation of the rostral cerebellar boundary. Development 125, 41–50 (1998).
Wijsman, E.M. et al. Genome-wide association of familial late-onset Alzheimer's disease replicates BIN1 and CLU and nominates CUGBP2 in interaction with APOE. PLoS Genet. 7, e1001308 (2011).
Lee, J.H., Cheng, R., Graff-Radford, N., Foroud, T. & Mayeux, R. Analyses of the National Institute on Aging Late-Onset Alzheimer's Disease Family Study: implication of additional loci. Arch. Neurol. 65, 1518–1526 (2008).
Nadeau, J.H. Modifier genes and protective alleles in humans and mice. Curr. Opin. Genet. Dev. 13, 290–295 (2003).
Goedert, M., Spillantini, M.G., Cairns, N.J. & Crowther, R.A. Tau proteins of Alzheimer paired helical filaments: abnormal phosphorylation of all six brain isoforms. Neuron 8, 159–168 (1992).
Glenner, G.G. & Wong, C.W. Alzheimer's disease: initial report of the purification and characterization of a novel cerebrovascular amyloid protein. Biochem. Biophys. Res. Commun. 120, 885–890 (1984).
Masters, C.L. et al. Neuronal origin of a cerebral amyloid: neurofibrillary tangles of Alzheimer's disease contain the same protein as the amyloid of plaque cores and blood vessels. EMBO J. 4, 2757–2763 (1985).
Llambi, F., Causeret, F., Bloch-Gallego, E. & Mehlen, P. Netrin-1 acts as a survival factor via its receptors UNC5H and DCC. EMBO J. 20, 2715–2722 (2001).
Thiebault, K. et al. The netrin-1 receptors UNC5H are putative tumor suppressors controlling cell death commitment. Proc. Natl. Acad. Sci. USA 100, 4173–4178 (2003).
Wang, R. et al. Autoinhibition of UNC5b revealed by the cytoplasmic domain structure of the receptor. Mol. Cell 33, 692–703 (2009).
Braak, H., Braak, E. & Bohl, J. Staging of Alzheimer-related cortical destruction. Eur. Neurol. 33, 403–408 (1993).
Ball, M.J. Neuronal loss, neurofibrillary tangles and granulovacuolar degeneration in the hippocampus with ageing and dementia. A quantitative study. Acta Neuropathol. 37, 111–118 (1977).
Squire, L.R. Memory and the hippocampus: a synthesis from findings with rats, monkeys, and humans. Psychol. Rev. 99, 195–231 (1992).
Jiao, B. et al. Investigation of TREM2, PLD3, and UNC5C variants in patients with Alzheimer's disease from mainland China. Neurobiol. Aging 35, 2422.e9–2422.e11 (2014).
Purcell, S. et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81, 559–575 (2007).
Liu, F., Kirichenko, A., Axenovich, T.I., van Duijn, C.M. & Aulchenko, Y.S. An approach for cutting large and complex pedigrees for linkage analysis. Eur. J. Hum. Genet. 16, 854–860 (2008).
Abecasis, G.R., Cherny, S.S., Cookson, W.O. & Cardon, L.R. Merlin–rapid analysis of dense genetic maps using sparse gene flow trees. Nat. Genet. 30, 97–101 (2002).
Drmanac, R. et al. Human genome sequencing using unchained base reads on self-assembling DNA nanoarrays. Science 327, 78–81 (2010).
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760 (2009).
DePristo, M.A. et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 43, 491–498 (2011).
Wang, K., Li, M. & Hakonarson, H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 38, e164 (2010).
Benitez, B.A. et al. TREM2 is associated with the risk of Alzheimer's disease in Spanish population. Neurobiol. Aging 34, 1711 e15–e17 (2013).
Cruchaga, C. et al. GWAS of cerebrospinal fluid tau levels identifies risk variants for Alzheimer's disease. Neuron 78, 256–268 (2013).
Jin, S.C. et al. Pooled-DNA sequencing identifies novel causative variants in PSEN1, GRN and MAPT in a clinical early-onset and familial Alzheimer's disease Ibero-American cohort. Alzheimers Res. Ther. 4, 34 (2012).
Braak, H. & Braak, E. Neuropathological stageing of Alzheimer-related changes. Acta Neuropathol. 82, 239–259 (1991).
Nagy, Z. et al. Assessment of the pathological stages of Alzheimer's disease in thin paraffin sections: a comparative study. Dement. Geriatr. Cogn. Disord. 9, 140–144 (1998).
Hyman, B.T. & Trojanowski, J.Q. Consensus recommendations for the postmortem diagnosis of Alzheimer disease from the National Institute on Aging and the Reagan Institute Working Group on diagnostic criteria for the neuropathological assessment of Alzheimer disease. J. Neuropathol. Exp. Neurol. 56, 1095–1097 (1997).
Price, A.L. et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 38, 904–909 (2006).
Nelson, C.D. & Sheng, M. Gpr3 stimulates Aβ production via interactions with APP and β-arrestin2. PLoS ONE 8, e74680 (2013).
Acknowledgements
This work was partially supported by grants from the US National Institutes of Health (R01-AG044546, P50-AG05681), and the Alzheimer's Association (NIRG-11-200110). This research was conducted while C.C. was a recipient of a New Investigator Award in Alzheimer's disease from the American Federation for Aging Research. C.C. is a recipient of a BrightFocus Foundation Alzheimer's Disease Research grant (A2013359S). Samples from the National Cell Repository for Alzheimer's Disease (NCRAD), which receives government support under a cooperative agreement grant (U24 AG21886) awarded by the National Institute on Aging (NIA), were used in this study. NIA-LOAD samples were collected under a cooperative agreement grant (U24 AG026395) awarded by the NIA. The ADGC is funded by the NIA (UO1AG032984). We thank K. Steffansøn and DeCode Genetics for genotype counts of UNC5C variants in the Icelandic population, the Alzheimer's Disease Centers, who collected samples used in this study, and patients and their families, whose help and participation made this work possible. We thank J. Norton of the Genetics Core, Washington University. We also thank C. Nelson (Genentech) for the HEK293 wild-type human APP695 stable cell line, K. Hoyte, Y. Lu, M. Sagolla, L. Gilmor, J. Borneo, E. Ladi, J. Grogan, J. Larson and J. Kaminker for technical assistance, and S. Ackerman, The Jackson Laboratory/Howard Hughes Medical Institute, for the generous gift of Unc5c−/− mice.
Author information
Authors and Affiliations
Consortia
Contributions
M.K.W.-S. and J. Hunkapiller designed, conducted and analyzed biological experiments and wrote the manuscript, T.R.B. performed linkage, case-control association, survival, growth-curve and sequencing data analysis, K.S. developed a method for isolating and sorting cell-type-specific adult brain for quantitative PCR, J.A.M. generated vectors and contributed to in vitro experiments, J.K.A. supervised the project, S.M.S. performed Unc5c in situ hybridization (ISH), M.B.Y. performed UNC5C ISH, O.F. contributed to ISH experiments and generated images, N.R. performed sample genotyping and handling and contributed to the manuscript, W.O. coordinated DNA sample collection, sequencing and data management, D.V.H. supervised development of the method for isolating and sorting cell-type-specific adult brain for quantitative PCR, M.T.-L. contributed to the manuscript, the ADGC provided AD case and control cohorts, R.M. provided sample material and contributed to the manuscript, M. P.-V., J. Haines, L.A.F., G.D.S. and A.G. provided sample material and contributed to the manuscript, T.W.B., C.C. and R.J.W. supervised the project and contributed to the manuscript, and R.R.G. supervised the project and wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
M.K.W.-S., J.H., T.R.B., K.S., J.A.M., J.K.A., S.M.S., M.B.Y., O.F., W.O., N.R., D.V.H., T.W.B., R.J.W. and R.R.G. are full-time employees of Genentech.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–14, Supplementary Tables 1–4 and Supplementary Results (PDF 131708 kb)
Live-cell imaging movies of cell death and caspase-3 or 7 activation in real time of β-gal-transfected HEK293 cells.
Representative live-cell imaging movie of β-gal-displaying transfected cells (red) and those cells with caspase-3 or 7 activity (green). (MOV 274 kb)
Live-cell imaging movies of cell death and caspase-3 or caspase-7 activation in real time of WT UNC5C-transfected HEK293 cells.
Representative live-cell imaging movie of WT UNC5C-displaying transfected cells (red) and those cells with caspase-3 or caspase-7 activity (green). (MOV 181 kb)
Live-cell imaging movies of cell death and caspase-3 or caspase-7 activation in real time of T835M UNC5C-transfected HEK293 cells.
Representative live-cell imaging movie of T835M UNC5C displaying transfected cells (red) and those cells with caspase-3 or 7 activity (green). (MOV 152 kb)
Live-cell imaging movies of β-gal-expressing hippocampal neurons.
Representative live-cell imaging movie of β-gal-expressing neurons exhibiting GFP+ transduced cells (green), as expression is initiated, maintained or lost as neurons undergo basal cell death. (MOV 2431 kb)
Live-cell imaging movies of WT UNC5C-expressing hippocampal neurons.
Representative live-cell imaging movie of WT UNC5C-expressing neurons exhibiting GFP+ transduced cells (green), as expression is initiated, maintained or lost as neurons undergo basal cell death. (MOV 2364 kb)
Live-cell imaging movies of T835M UNC5C-expressing hippocampal neurons undergoing basal cell death.
Representative live-cell imaging movie of T835M UNC5C-expressing neurons exhibiting GFP+ transduced cells (green), as expression is initiated, maintained or lost as neurons undergo basal cell death. (MOV 2328 kb)
Source data
Rights and permissions
About this article
Cite this article
Wetzel-Smith, M., Hunkapiller, J., Bhangale, T. et al. A rare mutation in UNC5C predisposes to late-onset Alzheimer's disease and increases neuronal cell death. Nat Med 20, 1452–1457 (2014). https://doi.org/10.1038/nm.3736
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nm.3736
This article is cited by
-
Death-associated protein kinase 1 as a therapeutic target for Alzheimer's disease
Translational Neurodegeneration (2024)
-
DCC/netrin-1 regulates cell death in oligodendrocytes after brain injury
Cell Death & Differentiation (2023)
-
Challenge accepted: uncovering the role of rare genetic variants in Alzheimer’s disease
Molecular Neurodegeneration (2022)
-
Associations of risk genes with onset age and plasma biomarkers of Alzheimer’s disease: a large case–control study in mainland China
Neuropsychopharmacology (2022)
-
Omics-based biomarkers discovery for Alzheimer's disease
Cellular and Molecular Life Sciences (2022)