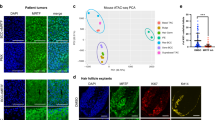
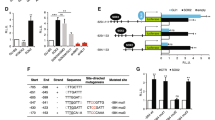
Abstract
Hedgehog signaling drives oncogenesis in several cancers, and strategies targeting this pathway have been developed, most notably through inhibition of Smoothened (SMO). However, resistance to Smoothened inhibitors occurs by genetic changes of Smoothened or other downstream Hedgehog components. Here we overcome these resistance mechanisms by modulating GLI transcription through inhibition of bromo and extra C-terminal (BET) bromodomain proteins. We show that BRD4 and other BET bromodomain proteins regulate GLI transcription downstream of SMO and suppressor of fused (SUFU), and chromatin immunoprecipitation studies reveal that BRD4 directly occupies GLI1 and GLI2 promoters, with a substantial decrease in engagement of these sites after treatment with JQ1, a small-molecule inhibitor targeting BRD4. Globally, genes associated with medulloblastoma-specific GLI1 binding sites are downregulated in response to JQ1 treatment, supporting direct regulation of GLI activity by BRD4. Notably, patient- and GEMM (genetically engineered mouse model)-derived Hedgehog-driven tumors (basal cell carcinoma, medulloblastoma and atypical teratoid rhabdoid tumor) respond to JQ1 even when harboring genetic lesions rendering them resistant to Smoothened antagonists. Altogether, our results reveal BET proteins as critical regulators of Hedgehog pathway transcriptional output and nominate BET bromodomain inhibitors as a strategy for treating Hedgehog-driven tumors with emerged or a priori resistance to Smoothened antagonists.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Accession codes
References
Goodrich, L.V., Johnson, R.L., Milenkovic, L., McMahon, J.A. & Scott, M.P. Conservation of the hedgehog/patched signaling pathway from flies to mice: induction of a mouse patched gene by Hedgehog. Genes Dev. 10, 301–312 (1996).
Binns, W., Shupe, J.L., Keeler, R.F. & James, L.F. Chronologic evaluation of teratogenicity in sheep fed Veratrum californicum. J. Am. Vet. Med. Assoc. 147, 839–842 (1965).
Gorlin, R.J., Vickers, R.A., Kellen, E. & Williamson, J.J. Multiple nasal-cell nevi syndrome. An analysis of a syndrome consisting of multiple nevoid basal-cell carcinoma, jaw cysts, skeletal anomalies, medulloblastoma, and hyporesponsiveness to parathormone. Cancer 18, 89–104 (1965).
Johnson, R.L. et al. Human homolog of patched, a candidate gene for the basal cell nevus syndrome. Science 272, 1668–1671 (1996).
Echelard, Y. et al. Sonic hedgehog, a member of a family of putative signaling molecules, is implicated in the regulation of CNS polarity. Cell 75, 1417–1430 (1993).
Marigo, V. et al. Cloning, expression, and chromosomal location of SHH and IHH: two human homologues of the Drosophila segment polarity gene hedgehog. Genomics 28, 44–51 (1995).
Alcedo, J., Ayzenzon, M., Von Ohlen, T., Noll, M. & Hooper, J.E. The Drosophila smoothened gene encodes a seven-pass membrane protein, a putative receptor for the hedgehog signal. Cell 86, 221–232 (1996).
Huangfu, D. & Anderson, K.V. Cilia and Hedgehog responsiveness in the mouse. Proc. Natl. Acad. Sci. USA 102, 11325–11330 (2005).
Haycraft, C.J. et al. Gli2 and Gli3 localize to cilia and require the intraflagellar transport protein polaris for processing and function. PLoS Genet. 1, e53 (2005).
Liu, A., Wang, B. & Niswander, L.A. Mouse intraflagellar transport proteins regulate both the activator and repressor functions of Gli transcription factors. Development 132, 3103–3111 (2005).
Oliver, T.G. et al. Transcriptional profiling of the Sonic hedgehog response: a critical role for N-myc in proliferation of neuronal precursors. Proc. Natl. Acad. Sci. USA 100, 7331–7336 (2003).
Regl, G. et al. Human GLI2 and GLI1 are part of a positive feedback mechanism in basal cell carcinoma. Oncogene 21, 5529–5539 (2002).
Mao, J. et al. A novel somatic mouse model to survey tumorigenic potential applied to the Hedgehog pathway. Cancer Res. 66, 10171–10178 (2006).
Xie, J. et al. Mutations of the PATCHED gene in several types of sporadic extracutaneous tumors. Cancer Res. 57, 2369–2372 (1997).
Thayer, S.P. et al. Hedgehog is an early and late mediator of pancreatic cancer tumorigenesis. Nature 425, 851–856 (2003).
Cho, Y.J. et al. Integrative genomic analysis of medulloblastoma identifies a molecular subgroup that drives poor clinical outcome. J. Clin. Oncol. 29, 1424–1430 (2011).
Pugh, T.J. et al. Medulloblastoma exome sequencing uncovers subtype-specific somatic mutations. Nature 488, 106–110 (2012).
Taylor, M.D. et al. Mutations in SUFU predispose to medulloblastoma. Nat. Genet. 31, 306–310 (2002).
ten Haaf, A. et al. Expression of the glioma-associated oncogene homolog (GLI) 1 in human breast cancer is associated with unfavourable overall survival. BMC Cancer 9, 298 (2009).
Northcott, P.A. et al. Subgroup-specific structural variation across 1,000 medulloblastoma genomes. Nature 488, 49–56 (2012).
Buczkowicz, P., Ma, J. & Hawkins, C. GLI2 is a potential therapeutic target in pediatric medulloblastoma. J. Neuropathol. Exp. Neurol. 70, 430–437 (2011).
Jagani, Z. et al. Loss of the tumor suppressor Snf5 leads to aberrant activation of the Hedgehog-Gli pathway. Nat. Med. 16, 1429–1433 (2010).
Zwerner, J.P. et al. The EWS/FLI1 oncogenic transcription factor deregulates GLI1. Oncogene 27, 3282–3291 (2008).
Chen, J.K., Taipale, J., Cooper, M.K. & Beachy, P.A. Inhibition of Hedgehog signaling by direct binding of cyclopamine to Smoothened. Genes Dev. 16, 2743–2748 (2002).
Buonamici, S. et al. Interfering with resistance to smoothened antagonists by inhibition of the PI3K pathway in medulloblastoma. Sci. Transl. Med. 2, 51ra70 (2010).
Lee, M.J. et al. Hedgehog pathway inhibitor saridegib (IPI-926) increases lifespan in a mouse medulloblastoma model. Proc. Natl. Acad. Sci. USA 109, 7859–7864 (2012).
Zhang, Y., Laterra, J. & Pomper, M.G. Hedgehog pathway inhibitor HhAntag691 is a potent inhibitor of ABCG2/BCRP and ABCB1/Pgp. Neoplasia 11, 96–101 (2009).
LoRusso, P.M. et al. Phase I trial of hedgehog pathway inhibitor vismodegib (GDC-0449) in patients with refractory, locally advanced or metastatic solid tumors. Clin. Cancer Res. 17, 2502–2511 (2011).
Rudin, C.M. et al. Treatment of medulloblastoma with hedgehog pathway inhibitor GDC-0449. N. Engl. J. Med. 361, 1173–1178 (2009).
Von Hoff, D.D. et al. Inhibition of the hedgehog pathway in advanced basal-cell carcinoma. N. Engl. J. Med. 361, 1164–1172 (2009).
Yauch, R.L. et al. Smoothened mutation confers resistance to a Hedgehog pathway inhibitor in medulloblastoma. Science 326, 572–574 (2009).
Kim, J. et al. Itraconazole and arsenic trioxide inhibit Hedgehog pathway activation and tumor growth associated with acquired resistance to smoothened antagonists. Cancer Cell 23, 23–34 (2013).
Kim, J., Lee, J.J., Gardner, D. & Beachy, P.A. Arsenic antagonizes the Hedgehog pathway by preventing ciliary accumulation and reducing stability of the Gli2 transcriptional effector. Proc. Natl. Acad. Sci. USA 107, 13432–13437 (2010).
Atwood, S.X., Li, M., Lee, A., Tang, J.Y. & Oro, A.E. GLI activation by atypical protein kinase C ιι/λ regulates the growth of basal cell carcinomas. Nature 494, 484–488 (2013).
Kenney, A.M., Widlund, H.R. & Rowitch, D.H. Hedgehog and PI-3 kinase signaling converge on Nmyc1 to promote cell cycle progression in cerebellar neuronal precursors. Development 131, 217–228 (2004).
Hyman, J.M. et al. Small-molecule inhibitors reveal multiple strategies for Hedgehog pathway blockade. Proc. Natl. Acad. Sci. USA 106, 14132–14137 (2009).
Filippakopoulos, P. et al. Selective inhibition of BET bromodomains. Nature 468, 1067–1073 (2010).
Dhalluin, C. et al. Structure and ligand of a histone acetyltransferase bromodomain. Nature 399, 491–496 (1999).
Jacobson, R.H., Ladurner, A.G., King, D.S. & Tjian, R. Structure and function of a human TAFII250 double bromodomain module. Science 288, 1422–1425 (2000).
Jang, M.K. et al. The bromodomain protein Brd4 is a positive regulatory component of P-TEFb and stimulates RNA polymerase II–dependent transcription. Mol. Cell 19, 523–534 (2005).
Yang, Z. et al. Recruitment of P-TEFb for stimulation of transcriptional elongation by the bromodomain protein Brd4. Mol. Cell 19, 535–545 (2005).
Whyte, W.A. et al. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell 153, 307–319 (2013).
Lovén, J. et al. Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell 153, 320–334 (2013).
Delmore, J.E. et al. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell 146, 904–917 (2011).
Mertz, J.A. et al. Targeting MYC dependence in cancer by inhibiting BET bromodomains. Proc. Natl. Acad. Sci. USA 108, 16669–16674 (2011).
Puissant, A. et al. Targeting MYCN in neuroblastoma by BET bromodomain inhibition. Cancer Discov. 3, 308–323 (2013).
Bandopadhayay, P. et al. BET bromodomain inhibition of MYC-amplified medulloblastoma. Clin. Cancer Res. 20, 912–925 (2014).
Chen, J.K., Taipale, J., Young, K.E., Maiti, T. & Beachy, P.A. Small molecule modulation of Smoothened activity. Proc. Natl. Acad. Sci. USA 99, 14071–14076 (2002).
Maity, T., Fuse, N. & Beachy, P.A. Molecular mechanisms of Sonic hedgehog mutant effects in holoprosencephaly. Proc. Natl. Acad. Sci. USA 102, 17026–17031 (2005).
Lewis, K.E., Concordet, J.P. & Ingham, P.W. Characterisation of a second patched gene in the zebrafish Danio rerio and the differential response of patched genes to Hedgehog signalling. Dev. Biol. 208, 14–29 (1999).
Shen, M.C. et al. Heat-shock–mediated conditional regulation of hedgehog/gli signaling in zebrafish. Dev. Dyn. 242, 539–549 (2013).
Macdonald, R. et al. Midline signalling is required for Pax gene regulation and patterning of the eyes. Development 121, 3267–3278 (1995).
Lee, J. et al. An ENU mutagenesis screen in zebrafish for visual system mutants identifies a novel splice-acceptor site mutation in patched2 that results in colobomas. Invest. Ophthalmol. Vis. Sci. 53, 8214–8221 (2012).
Chen, M.H. et al. Cilium-independent regulation of Gli protein function by Sufu in Hedgehog signaling is evolutionarily conserved. Genes Dev. 23, 1910–1928 (2009).
Roessler, E. et al. A previously unidentified amino-terminal domain regulates transcriptional activity of wild-type and disease-associated human GLI2. Hum. Mol. Genet. 14, 2181–2188 (2005).
Goodrich, L.V., Milenkovic, L., Higgins, K.M. & Scott, M.P. Altered neural cell fates and medulloblastoma in mouse patched mutants. Science 277, 1109–1113 (1997).
Aszterbaum, M. et al. Ultraviolet and ionizing radiation enhance the growth of BCCs and trichoblastomas in patched heterozygous knockout mice. Nat. Med. 5, 1285–1291 (1999).
Lee, E.Y. et al. Hedgehog pathway–regulated gene networks in cerebellum development and tumorigenesis. Proc. Natl. Acad. Sci. USA 107, 9736–9741 (2010).
Houzelstein, D. et al. Growth and early postimplantation defects in mice deficient for the bromodomain-containing protein Brd4. Mol. Cell. Biol. 22, 3794–3802 (2002).
Cooper, M.K., Porter, J.A., Young, K.E. & Beachy, P.A. Teratogen-mediated inhibition of target tissue response to Shh signaling. Science 280, 1603–1607 (1998).
Kool, M. et al. Genome sequencing of SHH medulloblastoma predicts genotype-related response to Smoothened inhibition. Cancer Cell 25, 393–405 (2014).
Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 102, 15545–15550 (2005).
Acknowledgements
This work was supported the St. Baldrick's Foundation Scholar Award (Y.-J.C.), a Beirne Faculty Scholar Endowment (Y.-J.C.), US National Institutes of Health (NIH) grant U01-CA176287 (Y.-J.C.), an Alex's Lemonade Stand Foundation Young Investigator Award (Y.T.), the Damon-Runyon Cancer Research Foundation (J.Q. and J.E.B.), NIH R01-CA159859 (R.W.-R.) and a pilot project grant from the Medical College of Wisconsin Cancer Center–Advancing a Healthier Wisconsin (B.A.L.). R.W.-R. is the recipient of a Leadership Award (LA1-01747) from the California Institute of Regenerative Medicine. We thank the Stanford Functional Genomics Facility (SFGF) and the Protein and Nucleic Acid (PAN) facility for their assistance in generating gene expression microarray data and reagents. We thank M. Scott (Stanford), C. Rudin (Memorial Sloan-Kettering Cancer Center), P. Beachy (Stanford), A. Sweet-Cordero (Stanford), P.-T. Chang (University of California, San Francisco), R. Karlstrom (University of Massachusetts, Amherst) and J.K. Chen (Stanford) for reagents, helpful suggestions and/or critical reading of the manuscript.
Author information
Authors and Affiliations
Contributions
Y.T. and Y.-J.C. conceived the project and wrote the manuscript. Y.T., B.N., S.M., B.B., N.V., S.S., S.C., A.P., S.O. and F.Y. performed all molecular biology experiments. J.Q. and J.E.B. synthesized and supplied JQ1 for all studies. A.E.O., S.X.A., R.J.W., A.L. and J.Y.T. generated and prepared GEMM-derived BCC cells, and R.W.-R. generated and provided patient-derived medulloblastoma cells. P.B., G.B., R.B. and Y.-J.C. performed all informatics analyses. S.G., A.L., Y.T., S.M., P.W., M.M., S.H.C. and S.S. performed JQ1 mouse in vivo studies. M.I.W. and B.A.L. performed all zebrafish studies.
Corresponding author
Ethics declarations
Competing interests
The Dana-Farber Cancer Institute has licensed drug-like derivatives of JQ1 prepared in the Bradner laboratory to Tensha Therapeutics for clinical translation as cancer therapeutics. Dana-Farber and J.E.B. have been provided minority equity shares in Tensha. J.Q. has a consultant agreement with Tensha Therapeutics.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–7 (PDF 3270 kb)
Supplementary Data Set
ssGSEA of gene expression data generated from SmoWT MB cells exposed for 6h to DMSO, JQ1 (1 μM), or GDC-0449 (0.1 μM) using v2.0 CGP gene sets (http://www.broadinstitute.org/gsea/msigdb/index.jsp). (TXT 830 kb)
Supplementary Table 1
Gene sets derived from Lee et al., PNAS 2010. (XLSX 23 kb)
Supplementary Table 2
qPCR primers used in study. (XLSX 9 kb)
Rights and permissions
About this article
Cite this article
Tang, Y., Gholamin, S., Schubert, S. et al. Epigenetic targeting of Hedgehog pathway transcriptional output through BET bromodomain inhibition. Nat Med 20, 732–740 (2014). https://doi.org/10.1038/nm.3613
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nm.3613
This article is cited by
-
SALL4 is a CRL3REN/KCTD11 substrate that drives Sonic Hedgehog-dependent medulloblastoma
Cell Death & Differentiation (2024)
-
Targeted protein degradation reveals BET bromodomains as the cellular target of Hedgehog pathway inhibitor-1
Nature Communications (2023)
-
New cytotoxic dammarane type saponins from Ziziphus spina-christi
Scientific Reports (2023)
-
Therapeutic targeting the oncogenic driver EWSR1::FLI1 in Ewing sarcoma through inhibition of the FACT complex
Oncogene (2023)
-
Bromodomain and extraterminal (BET) proteins: biological functions, diseases, and targeted therapy
Signal Transduction and Targeted Therapy (2023)