Abstract
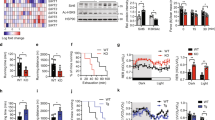
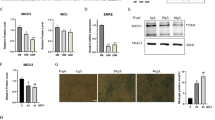
The nuclear receptor Rev-erb-α modulates hepatic lipid and glucose metabolism, adipogenesis and the inflammatory response in macrophages. We show here that Rev-erb-α is highly expressed in oxidative skeletal muscle and that its deficiency in muscle leads to reduced mitochondrial content and oxidative function, as well as upregulation of autophagy. These cellular effects resulted in both impaired mitochondrial biogenesis and increased clearance of this organelle, leading to compromised exercise capacity. On a molecular level, Rev-erb-α deficiency resulted in deactivation of the Lkb1-Ampk-Sirt1–Ppargc-1α signaling pathway. These effects were recapitulated in isolated fibers and in muscle cells after knockdown of the gene encoding Rev-erb-α, Nr1d1. In complementary experiments, Rev-erb-α overexpression in vitro increased the number of mitochondria and improved respiratory capacity, whereas muscle overexpression or pharmacological activation of Rev-erb-α in vivo increased exercise capacity. This study identifies Rev-erb-α as a pharmacological target that improves muscle oxidative function by modulating gene networks controlling mitochondrial number and function.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Lin, J. et al. Transcriptional co-activator PGC-1 α drives the formation of slow-twitch muscle fibres. Nature 418, 797–801 (2002).
Schuler, M. et al. PGC1α expression is controlled in skeletal muscles by PPARβ, whose ablation results in fiber-type switching, obesity, and type 2 diabetes. Cell Metab. 4, 407–414 (2006).
Narkar, V.A. et al. Exercise and PGC-1α-independent synchronization of type I muscle metabolism and vasculature by ERRgamma. Cell Metab. 13, 283–293 (2011).
Zechner, C. et al. Total skeletal muscle PGC-1 deficiency uncouples mitochondrial derangements from fiber type determination and insulin sensitivity. Cell Metab. 12, 633–642 (2010).
Yamamoto, H. et al. NCoR1 is a conserved physiological modulator of muscle mass and oxidative function. Cell 147, 827–839 (2011).
Handschin, C. et al. Skeletal muscle fiber-type switching, exercise intolerance, and myopathy in PGC-1α muscle-specific knock-out animals. J. Biol. Chem. 282, 30014–30021 (2007).
Leone, T.C. et al. PGC-1α deficiency causes multi-system energy metabolic derangements: muscle dysfunction, abnormal weight control and hepatic steatosis. PLoS Biol. 3, e101 (2005).
Duez, H. & Staels, B. Nuclear receptors linking circadian rhythms and cardiometabolic control. Arterioscler. Thromb. Vasc. Biol. 30, 1529–1534 (2010).
Duez, H. et al. Regulation of bile acid synthesis by the nuclear receptor Rev-erbα. Gastroenterology 135, 689–698 (2008).
Yin, L. et al. Rev-erbα, a heme sensor that coordinates metabolic and circadian pathways. Science 318, 1786–1789 (2007).
Cho, H. et al. Regulation of circadian behaviour and metabolism by REV-ERB-α and REV-ERB-β. Nature 485, 123–127 (2012).
Solt, L.A. et al. Regulation of circadian behaviour and metabolism by synthetic REV-ERB agonists. Nature 485, 62–68 (2012).
Bugge, A. et al. Rev-erbα and Rev-erbβ coordinately protect the circadian clock and normal metabolic function. Genes Dev. 26, 657–667 (2012).
Fontaine, C. et al. The orphan nuclear receptor Rev-Erbα is a peroxisome proliferator-activated receptor (PPAR) γ target gene and promotes PPARγ-induced adipocyte differentiation. J. Biol. Chem. 278, 37672–37680 (2003).
Wang, J. & Lazar, M.A. Bifunctional role of Rev-erbα in adipocyte differentiation. Mol. Cell Biol. 28, 2213–2220 (2008).
Fontaine, C. et al. The nuclear receptor Rev-erbα is a liver X receptor (LXR) target gene driving a negative feedback loop on select LXR-induced pathways in human macrophages. Mol. Endocrinol. 22, 1797–1811 (2008).
Feng, D. et al. A circadian rhythm orchestrated by histone deacetylase 3 controls hepatic lipid metabolism. Science 331, 1315–1319 (2011).
Wu, N., Yin, L., Hanniman, E.A., Joshi, S. & Lazar, M.A. Negative feedback maintenance of heme homeostasis by its receptor, Rev-erbα. Genes Dev. 23, 2201–2209 (2009).
Estall, J.L. et al. PGC-1α negatively regulates hepatic FGF21 expression by modulating the heme/Rev-Erbα axis. Proc. Natl. Acad. Sci. USA 106, 22510–22515 (2009).
Cantó, C. et al. Interdependence of AMPK and SIRT1 for metabolic adaptation to fasting and exercise in skeletal muscle. Cell Metab. 11, 213–219 (2010).
Narendra, D., Tanaka, A., Suen, D.F. & Youle, R.J. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J. Cell Biol. 183, 795–803 (2008).
Asp, P. et al. Genome-wide remodeling of the epigenetic landscape during myogenic differentiation. Proc. Natl. Acad. Sci. 108, E149–E158 (2011).
Fujii, N. et al. Role of AMP-activated protein kinase in exercise capacity, whole body glucose homeostasis, and glucose transport in skeletal muscle -insight from analysis of a transgenic mouse model. Diabetes Res. Clin. Pract. 77 (suppl. 1), S92–S98 (2007).
Koh, H.J. et al. Skeletal muscle-selective knockout of LKB1 increases insulin sensitivity, improves glucose homeostasis, and decreases TRB3. Mol. Cell Biol. 26, 8217–8227 (2006).
Thomson, D.M. et al. Skeletal muscle dysfunction in muscle-specific LKB1 knockout mice. J. Appl. Physiol. 108, 1775–1785 (2010).
Mihaylova, M.M. & Shaw, R.J. The AMPK signalling pathway coordinates cell growth, autophagy and metabolism. Nat. Cell Biol. 13, 1016–1023 (2011).
Rabinowitz, J.D. & White, E. Autophagy and metabolism. Science 330, 1344–1348 (2010).
Kroemer, G., Marino, G. & Levine, B. Autophagy and the integrated stress response. Mol. Cell 40, 280–293 (2010).
Kabeya, Y. et al. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 19, 5720–5728 (2000).
Joo, J.H. et al. Hsp90-Cdc37 chaperone complex regulates Ulk1- and Atg13-mediated mitophagy. Mol. Cell 43, 572–585 (2011).
Kundu, M. et al. Ulk1 plays a critical role in the autophagic clearance of mitochondria and ribosomes during reticulocyte maturation. Blood 112, 1493–1502 (2008).
Egan, D.F. et al. Phosphorylation of ULK1 (hATG1) by AMP-activated protein kinase connects energy sensing to mitophagy. Science 331, 456–461 (2011).
Lee, I.H. et al. A role for the NAD-dependent deacetylase Sirt1 in the regulation of autophagy. Proc. Natl. Acad. Sci. USA 105, 3374–3379 (2008).
Bass, J. Circadian topology of metabolism. Nature 491, 348–356 (2012).
McCarthy, J.J. et al. Identification of the circadian transcriptome in adult mouse skeletal muscle. Physiol. Genomics 31, 86–95 (2007).
Miller, B.H. et al. Circadian and CLOCK-controlled regulation of the mouse transcriptome and cell proliferation. Proc. Natl. Acad. Sci. USA 104, 3342–3347 (2007).
Andrews, J.L. et al. CLOCK and BMAL1 regulate MyoD and are necessary for maintenance of skeletal muscle phenotype and function. Proc. Natl. Acad. Sci. USA 107, 19090–19095 (2010).
Duez, H. et al. Regulation of bile acid synthesis by the nuclear receptor Rev-erbalpha. Gastroenterology 135, 689–698 (2008).
Nagoshi, E. et al. Circadian gene expression in individual fibroblasts: cell-autonomous and self-sustained oscillators pass time to daughter cells. Cell 119, 693–705 (2004).
Raspé, E. et al. Identification of Rev-erbα as a physiological repressor of apoC-III gene transcription. J. Lipid Res. 43, 2172–2179 (2002).
Aragonés, J. et al. Deficency or inhibition of oxygen sensor Phd1 induces hypoxia tolerance by reprogramming basal metabolism. Nat. Genet. 40, 170–180 (2008).
Chomczynski, P. & Sacchi, N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 162, 156–159 (1987).
Delhaas, T. et al. Steep increase in myonuclear domain size during infancy. Anat. Rec. 296, 192–197 (2013).
Kaminskyy, V. et al. A quantitative assay for the monitoring of autophagosome accumulation in different phases of the cell cycle. Autophagy 7, 83–90 (2011).
Eeckhoute, J., Lupien, M. & Brown, M. Combining chromatin immunoprecipitation and oligonucleotide tiling arrays (ChIP-Chip) for functional genomic studies. Methods Mol. Biol. 556, 155–164 (2009).
Acknowledgements
This research was supported by a Marie Curie International Reintegration Grant (FP7) (to H.D.), the European Commission (FP7) consortium Eurhythdia (to B.S.), Région Nord Pas-de-Calais/Fonds Européen de Développement Régional (to B.S.), a Contrat de Projet Etat-Région 'starting grant' (to H.D.), the European Genomic Institute for Diabetes (EGID, ANR-10-LABX-46) (to B.S.), an unrestricted Instituts Thématiques Multi-Organismes/Astra Zeneca grant (to B.S.), a joint Société Francophone du Diabète/Merck Sharp & Dohme research fellowship (to H.D.), a research grant from the European Foundation for the Study of Diabetes/Lilly (to H.D.), US National Institutes of Health grants (MH093429 and DK080201) (to T.P.B.), a National Research Service Award (DK088499) (to L.A.S.) and a VICI research grant for innovative research from the Netherlands Organization for Scientific Research (918.96.618) (to P.S.). B.S. receives support from the Institut Universitaire de France.
Author information
Authors and Affiliations
Contributions
E.W., Y. Sebti, B.S. and H.D. were responsible for the study design, data analysis and interpretation and wrote the manuscript. E.W., Y. Sebti, L.A.S., C.D., S.L., C.P., S.D., G.S. and R.N. performed the experiments and data analysis. J.E., P.L., M.K.C.H., P.S. and T.P.B. were involved in data analysis. Y. Shin and T.M.K. were involved in Rev-erb-α ligand chemistry.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–8 and Supplementary Tables 1–2 (PDF 542 kb)
Rights and permissions
About this article
Cite this article
Woldt, E., Sebti, Y., Solt, L. et al. Rev-erb-α modulates skeletal muscle oxidative capacity by regulating mitochondrial biogenesis and autophagy. Nat Med 19, 1039–1046 (2013). https://doi.org/10.1038/nm.3213
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nm.3213
This article is cited by
-
Importance of Per2 in cardiac mitochondrial protection during stress
Scientific Reports (2024)
-
Targeting NR1D1 in organ injury: challenges and prospects
Military Medical Research (2023)
-
Brain nuclear receptors and cardiovascular function
Cell & Bioscience (2023)
-
Impact of the circadian nuclear receptor REV-ERBα in dorsal raphe 5-HT neurons on social interaction behavior, especially social preference
Experimental & Molecular Medicine (2023)
-
An electroaffinity labelling platform for chemoproteomic-based target identification
Nature Chemistry (2023)