Abstract
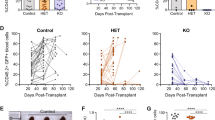
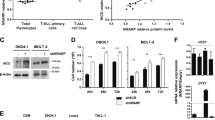
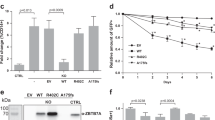
Reactive oxygen species (ROS), a byproduct of cellular metabolism, damage intracellular macromolecules and, when present in excess, can promote normal hematopoietic stem cell differentiation and exhaustion1,2,3. However, mechanisms that regulate the amount of ROS in leukemia-initiating cells (LICs) and the biological role of ROS in these cells are largely unknown. We show here that the ROSlow subset of CD44+ cells in T cell acute lymphoblastic leukemia (T-ALL), a malignancy of immature T cell progenitors, is highly enriched in the most aggressive LICs and that ROS accumulation is restrained by downregulation of protein kinase C θ (PKC-θ). Notably, primary mouse T-ALLs lacking PKC-θ show improved LIC activity, whereas enforced PKC-θ expression in both mouse and human primary T-ALLs compromised LIC activity. We also show that PKC-θ is regulated by a new pathway in which NOTCH1 induces runt-related transcription factor 3 (RUNX3), RUNX3 represses RUNX1 and RUNX1 induces PKC-θ. NOTCH1, which is frequently activated by mutation in T-ALL4,5,6 and required for LIC activity in both mouse and human models7,8, thus acts to repress PKC-θ. These results reveal key functional roles for PKC-θ and ROS in T-ALL and suggest that aggressive biological behavior in vivo could be limited by therapeutic strategies that promote PKC-θ expression or activity, or the accumulation of ROS.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Accession codes
References
Ito, K. et al. Reactive oxygen species act through p38 MAPK to limit the lifespan of hematopoietic stem cells. Nat. Med. 12, 446–451 (2006).
Owusu-Ansah, E. & Banerjee, U. Reactive oxygen species prime Drosophila haematopoietic progenitors for differentiation. Nature 461, 537–541 (2009).
Tothova, Z. et al. FoxOs are critical mediators of hematopoietic stem cell resistance to physiologic oxidative stress. Cell 128, 325–339 (2007).
O′Neil, J. et al. FBW7 mutations in leukemic cells mediate NOTCH pathway activation and resistance to γ-secretase inhibitors. J. Exp. Med. 204, 1813–1824 (2007).
Thompson, B.J. et al. The SCFFBW7 ubiquitin ligase complex as a tumor suppressor in T cell leukemia. J. Exp. Med. 204, 1825–1835 (2007).
Weng, A.P. et al. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science 306, 269–271 (2004).
Armstrong, F. et al. NOTCH is a key regulator of human T-cell acute leukemia initiating cell activity. Blood 113, 1730–1740 (2009).
Tatarek, J. et al. Notch1 inhibition targets the leukemia-initiating cells in a Tal1/Lmo2 mouse model of T-ALL. Blood 118, 1579–1590 (2011).
Pui, C.H. & Evans, W.E. Treatment of acute lymphoblastic leukemia. N. Engl. J. Med. 354, 166–178 (2006).
Cox, C.V. et al. Characterization of a progenitor cell population in childhood T-cell acute lymphoblastic leukemia. Blood 109, 674–682 (2007).
Gerby, B. et al. Expression of CD34 and CD7 on human T-cell acute lymphoblastic leukemia discriminates functionally heterogeneous cell populations. Leukemia 25, 1249–1258 (2011).
Chiu, P.P., Jiang, H. & Dick, J.E. Leukemia-initiating cells in human T-lymphoblastic leukemia exhibit glucocorticoid resistance. Blood 116, 5268–5279 (2010).
Reya, T., Morrison, S.J., Clarke, M.F. & Weissman, I.L. Stem cells, cancer, and cancer stem cells. Nature 414, 105–111 (2001).
Kobayashi, C.I. & Suda, T. Regulation of reactive oxygen species in stem cells and cancer stem cells. J. Cell. Physiol. 227, 421–430 (2012).
Valko, M. et al. Free radicals and antioxidants in normal physiological functions and human disease. Int. J. Biochem. Cell Biol. 39, 44–84 (2007).
Adly, A.A.M. Oxidative stress and disease: an updated review. Res. J. Immunol. 3, 129–145 (2010).
Guzman, M.L. et al. The sesquiterpene lactone parthenolide induces apoptosis of human acute myelogenous leukemia stem and progenitor cells. Blood 105, 4163–4169 (2005).
Diehn, M. et al. Association of reactive oxygen species levels and radioresistance in cancer stem cells. Nature 458, 780–783 (2009).
Chiang, M.Y. et al. Leukemia-associated NOTCH1 alleles are weak tumor initiators but accelerate K-ras–initiated leukemia. J. Clin. Invest. 118, 3181–3194 (2008).
Medyouf, H. et al. Acute T-cell leukemias remain dependent on Notch signaling despite PTEN and INK4A/ARF loss. Blood 115, 1175–1184 (2010).
Pear, W.S. et al. Exclusive development of T cell neoplasms in mice transplanted with bone marrow expressing activated Notch alleles. J. Exp. Med. 183, 2283–2291 (1996).
Eruslanov, E. & Kusmartsev, S. Identification of ROS using oxidized DCFDA and flow-cytometry. Methods Mol. Biol. 594, 57–72 (2010).
Felli, M.P. et al. PKCθ mediates pre-TCR signaling and contributes to Notch3-induced T-cell leukemia. Oncogene 24, 992–1000 (2005).
Sun, Z. et al. PKC-θ is required for TCR-induced NF-κB activation in mature but not immature T lymphocytes. Nature 404, 402–407 (2000).
Kaminski, M., Kiessling, M., Suss, D., Krammer, P.H. & Gulow, K. Novel role for mitochondria: protein kinase Cθ-dependent oxidative signaling organelles in activation-induced T-cell death. Mol. Cell. Biol. 27, 3625–3639 (2007).
Baier-Bitterlich, G. et al. Protein kinase C-θ isoenzyme selective stimulation of the transcription factor complex AP-1 in T lymphocytes. Mol. Cell. Biol. 16, 1842–1850 (1996).
Della Gatta, G. et al. Reverse engineering of TLX oncogenic transcriptional networks identifies RUNX1 as tumor suppressor in T-ALL. Nat. Med. 18, 436–440 (2012).
Grossmann, V. et al. Prognostic relevance of RUNX1 mutations in T-cell acute lymphoblastic leukemia. Haematologica 96, 1874–1877 (2011).
Zhang, J. et al. The genetic basis of early T-cell precursor acute lymphoblastic leukaemia. Nature 481, 157–163 (2012).
Coustan-Smith, E. et al. Early T-cell precursor leukaemia: a subtype of very high-risk acute lymphoblastic leukaemia. Lancet Oncol. 10, 147–156 (2009).
Gutierrez, A. et al. The BCL11B tumor suppressor is mutated across the major molecular subtypes of T-cell acute lymphoblastic leukemia. Blood 118, 4169–4173 (2011).
Homminga, I. et al. Integrated transcript and genome analyses reveal NKX2-1 and MEF2C as potential oncogenes in T cell acute lymphoblastic leukemia. Cancer Cell 19, 484–497 (2011).
Winter, S.S. et al. Identification of genomic classifiers that distinguish induction failure in T-lineage acute lymphoblastic leukemia: a report from the Children′s Oncology Group. Blood 110, 1429–1438 (2007).
Heintzman, N.D. et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat. Genet. 39, 311–318 (2007).
Spender, L.C., Whiteman, H.J., Karstegl, C.E. & Farrell, P.J. Transcriptional cross-regulation of RUNX1 by RUNX3 in human B cells. Oncogene 24, 1873–1881 (2005).
Wang, H. et al. Genome-wide analysis reveals conserved and divergent features of Notch1/RBPJ binding in human and murine T-lymphoblastic leukemia cells. PNAS 108, 14908–14913 (2011).
Weng, A.P. et al. Growth suppression of pre-T acute lymphoblastic leukemia cells by inhibition of notch signaling. Mol. Cell. Biol. 23, 655–664 (2003).
Guo, W. et al. Multi-genetic events collaboratively contribute to Pten-null leukaemia stem-cell formation. Nature 453, 529–533 (2008).
Tremblay, M. et al. Modeling T-cell acute lymphoblastic leukemia induced by the SCL and LMO1 oncogenes. Genes Dev 24, 1093–1105 (2010).
Isakov, N. & Altman, A. Protein kinase Cθ in T cell activation. Annu. Rev. Immunol. 20, 761–794 (2002).
Morita, S., Kojima, T. & Kitamura, T. Plat-E: an efficient and stable system for transient packaging of retroviruses. Gene Ther. 7, 1063–1066 (2000).
Zufferey, R., Nagy, D., Mandel, R.J., Naldini, L. & Trono, D. Multiply attenuated lentiviral vector achieves efficient gene delivery in vivo. Nat. Biotechnol. 15, 871–875 (1997).
Dull, T. et al. A third-generation lentivirus vector with a conditional packaging system. J. Virol. 72, 8463–8471 (1998).
Zufferey, R. et al. Self-inactivating lentivirus vector for safe and efficient in vivo gene delivery. J. Virol. 72, 9873–9880 (1998).
Zennou, V. et al. HIV-1 genome nuclear import is mediated by a central DNA flap. Cell 101, 173–185 (2000).
Sirven, A. et al. The human immunodeficiency virus type-1 central DNA flap is a crucial determinant for lentiviral vector nuclear import and gene transduction of human hematopoietic stem cells. Blood 96, 4103–4110 (2000).
Logan, A.C. et al. Factors influencing the titer and infectivity of lentiviral vectors. Hum. Gene Ther. 15, 976–988 (2004).
Robbins, P.B. et al. Consistent, persistent expression from modified retroviral vectors in murine hematopoietic stem cells. Proc. Natl. Acad. Sci. USA 95, 10182–10187 (1998).
Root, D.E., Hacohen, N., Hahn, W.C., Lander, E.S. & Sabatini, D.M. Genome-scale loss-of-function screening with a lentiviral RNAi library. Nat. Methods 3, 715–719 (2006).
Ou, W.B., Zhu, M.J., Demetri, G.D., Fletcher, C.D.M. & Fletcher, J.A. Protein kinase C-θ regulates KIT expression and proliferation in gastrointestinal stromal tumors. Oncogene 27, 5624–5634 (2008).
Sarbassov, D.D., Guertin, D.A., Ali, S.M. & Sabatini, D.M. Phosphorylation and Regulation of Akt/PKB by the Rictor-mTOR complex. Science 307, 1098–1101 (2005).
Palomero, T. et al. Mutational loss of PTEN induces resistance to NOTCH1 inhibition in T-cell leukemia. Nat. Med. 13, 1203–1210 (2007).
Palomero, T. et al. CUTLL1, a novel human T-cell lymphoma cell line with t(7;9) rearrangement, aberrant NOTCH1 activation and high sensitivity to γ-secretase inhibitors. Leukemia 20, 1279–1287 (2006).
Dallas, M.H., Varnum-Finney, B., Martin, P.J. & Bernstein, I.D. Enhanced T-cell reconstitution by hematopoietic progenitors expanded ex vivo using the Notch ligand Δ1. Blood 109, 3579–3587 (2007).
Schneider, C.A., Rasband, W.S. & Eliceiri, K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675 (2012).
Hasserjian, R.P., Aster, J.C., Davi, F., Weinberg, D.S. & Sklar, J. Modulated expression of notch1 during thymocyte development. Blood 88, 970–976 (1996).
Ji, H. et al. An integrated software system for analyzing ChIP-chip and ChIP-seq data. Nat. Biotechnol. 26, 1293–1300 (2008).
Li, C. & Wong, W.H. Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. PNAS 98, 31–36 (2001).
Hu, Y. & Smyth, G.K. ELDA: extreme limiting dilution analysis for comparing depleted and enriched populations in stem cell and other assays. J. Immunol. Methods 347, 70–78 (2009).
Acknowledgements
We would like to thank Z. Sun (City of Hope, Duarte, California) for providing PKC-θ knockout mice, J. Fletcher and W. Ou (Brigham and Women's Hospital, Boston, Massachusetts) for PKC-θ shRNA lentivectors, E. Kieff (Brigham and Women's Hospital, Boston, Massachusetts) for CSL antibody, I. Bernstein (Fred Hutchinson Cancer Research Center, Seattle, Washington) for Ig-DL1 ligand, F. Pflumio (Commissariat à l'Énergie Atomique Fontenay-aux-Roses, France) for the MS5-DL1 cell line and E. Martignani for help with immunofluorescence. We would also like to thank P. Ballerini (Hôpital Armand Trousseau, Paris, France), T. Leblanc (Hôpital Saint-Louis, Paris, France), L. Matherly (Karmanos Cancer Institute, Detroit, Michigan) and M.J. You (MD Anderson Cancer Center, Houston, Texas) for providing patient T-ALL samples. This work was supported by grants from the Canadian Institutes of Health Research/Terry Fox Foundation, the Leukemia and Lymphoma Society of Canada, the Cancer Research Society, the Lymphoma Foundation Canada (to A.P.W.) and the US National Cancer Institute (P01CA119070 to J.C.A.). M.Y.C. was supported by a career development award from the US National Cancer Institute (K08CA120544). A.P.W. was a Michael Smith Foundation for Health Research Scholar.
Author information
Authors and Affiliations
Contributions
V.G. and A.P.W. designed the experiments. V.G., C.R.J., S.H.L., O.O.S., O.N., C.W. and S.G. generated the data. V.G. and A.P.W. interpreted the results. M.Y.C. provided reagents and advice. H.W. and J.C.A. generated and analyzed ChIP-Seq data. R.K.H. and C.E. provided advice and discussion. V.G. and A.P.W. wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–15 and Supplementary Table 1 (PDF 1576 kb)
Rights and permissions
About this article
Cite this article
Giambra, V., Jenkins, C., Wang, H. et al. NOTCH1 promotes T cell leukemia-initiating activity by RUNX-mediated regulation of PKC-θ and reactive oxygen species. Nat Med 18, 1693–1698 (2012). https://doi.org/10.1038/nm.2960
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nm.2960
This article is cited by
-
The circadian clock circuitry modulates leukemia initiating cell activity in T-cell acute lymphoblastic leukemia
Journal of Experimental & Clinical Cancer Research (2023)
-
PD-1 signalling defines and protects leukaemic stem cells from T cell receptor-induced cell death in T cell acute lymphoblastic leukaemia
Nature Cell Biology (2023)
-
Targeting the NOTCH1-MYC-CD44 axis in leukemia-initiating cells in T-ALL
Leukemia (2022)
-
Redox balance and autophagy regulation in cancer progression and their therapeutic perspective
Medical Oncology (2022)
-
Macrophage mitochondrial fission improves cancer cell phagocytosis induced by therapeutic antibodies and is impaired by glutamine competition
Nature Cancer (2022)