Abstract
Protective immunity against influenza virus infection is mediated by neutralizing antibodies, but the precise role of T cells in human influenza immunity is uncertain. We conducted influenza infection studies in healthy volunteers with no detectable antibodies to the challenge viruses H3N2 or H1N1. We mapped T cell responses to influenza before and during infection. We found a large increase in influenza-specific T cell responses by day 7, when virus was completely cleared from nasal samples and serum antibodies were still undetectable. Preexisting CD4+, but not CD8+, T cells responding to influenza internal proteins were associated with lower virus shedding and less severe illness. These CD4+ cells also responded to pandemic H1N1 (A/CA/07/2009) peptides and showed evidence of cytotoxic activity. These cells are an important statistical correlate of homotypic and heterotypic response and may limit severity of influenza infection by new strains in the absence of specific antibody responses. Our results provide information that may aid the design of future vaccines against emerging influenza strains.
Similar content being viewed by others
Main
Influenza viruses evade established protective immune responses by two distinct mechanisms: gradual antigenic drift of viral surface epitopes resulting from low-fidelity viral replication, which allows escape from established serological immunity1,2, and the emergence of entirely new viral strains arising from the reassortment of influenza virus RNA from different strains in a common host3,4. The emerging new pathogen can result in pandemics, which spread through communities that lack protective immunity to the previously unencountered viral proteins.
Influenza infection studies in mice have revealed that viral clearance is mediated by antigen-specific CD8+ effector T cells, whereas memory CD4+ T cells are important in maintaining CD8+ T and B cell memory responses5. Recently, both effector CD4+ and CD8+ T cells have been implicated in the control of pulmonary inflammation and limit excessive tissue damage by producing interleukin-10 (ref. 6).
In the context of pandemics where there are no preexisting protective antibodies, T cells may mediate protection or limit the severity of influenza-associated illness in humans7. Preexisting T cell responses have been shown to modulate influenza severity in the context of existing antibodies8, but the role of protective cell-mediated immunity (CMI) in seronegative individuals naive to a particular strain is not understood.
Here we investigate the role of CMI in limiting influenza using a human challenge model in healthy volunteers who lack detectable humoral immunity to the challenge strains. We focus on responses to peptides from the internal influenza proteins, which are highly conserved across strains, and identify a role for CD4+ T cells recognizing these proteins in limiting disease severity. By mapping the exact nature of the epitopes generating these responses across strains, we provide direction for the development of broadly protective T cell vaccines.
Results
Human influenza infection model
We established an experimental infection model using live influenza A virus in human volunteers9. Forty-one healthy volunteers aged between 18 and 45 were inoculated intranasally with serial tenfold dilutions of influenza A viruses: a cell-grown H3N2 WS/67/05 or an egg-grown H1N1 BR/59/07. We studied subjects in a clinical isolation facility and measured viral shedding, symptom development and cellular and humoral immune responses for the first 7 d and at day 28. In the H3N2 challenge, 8 out of 17 (47%) volunteers were female, and the median age was 26.5 years (range 22–41) (Supplementary Table 1). In the H1N1 study, 7 out of 24 (29%) were female, and the median age was 24 years (range 19–35).
All volunteers selected were seronegative to the challenge strain and PCR negative for the virus in nasal lavage. We defined the overall infection rate by evidence of virus shedding or seroconversion by day 28. The infection rate was higher in subjects challenged with H3N2 (14/17, 82%) than in subjects challenged with H1N1 (9/24, 38%).
In the H3N2 challenge, virus shedding in nasal lavage persisted in individuals for as long as 7 d, but most subjects (8/14, 57%) cleared the virus completely by day 4 (Fig. 1a). The H1N1 group did not show high levels of viral shedding in nasal lavage of all infected individuals, a phenomenon that has been described previously with egg-grown influenza virus when first passaged through mammalian cells10.
(a) Virus in nasal samples of volunteers infected with cell-grown H3N2 (WS/67/05) virus or egg-grown H1N1 (BR/59/07) virus, as titrated by half-maximal tissue culture infectious dose (TCID50) assay. Error bars represent s.e.m. (b) Correlation of total symptom scores against the peak nasal virus shedding in H3N2-infected subjects by Spearman rank correlation test. (c) Mean symptom scores and oral temperatures of volunteers infected with H3N2 virus. (d) Mean symptom scores and oral temperatures of volunteers infected with H1N1 virus. Symptom assessments were performed by the volunteers twice daily on a four-point scale (absent to severe). The score for each symptom group was obtained by adding the total individual symptom scores for that particular group on that particular day. Oral temperatures were determined four times a day for the duration of the study, and the highest temperature was represented.
In the H3N2 infection, total symptoms tracked with peak viral load (Fig. 1b, r = 0.6977, P = 0.0055, Spearman coefficient). Similar symptom profiles were observed between the two cohorts and were comparable to natural infections11. In the H3N2 group, 11/14 (79%) infected subjects developed one or more symptoms and as a group had symptom scores that peaked on day 3 and returned to normal by day 7 after inoculation (Fig. 1c). Three out of 14 subjects (21%) developed fever (oral temperature > 37.7 °C), and the highest temperatures were detected on day 2. In the H1N1 group, 8/9 (89%) infected subjects developed one or more symptoms, and mean symptom scores peaked on day 4 and returned to normal by day 7 after viral inoculation (Fig. 1d). One out of nine infected subjects (11%) developed fever, and the highest temperatures were detected on day 2.
In both groups, upper respiratory symptoms predominated; runny nose and sore throat occurred in 10/14 (71%) subjects in the H3N2 group and 8/9 (89%) subjects in the H1N1 group. Lower respiratory symptoms were much milder in severity and occurred in 3/14 (21%) individuals in the H3N2 group and 2/9 (22%) individuals in the H1N1 group. Systemic symptoms such as muscle aches were also present in 6/14 (43%) subjects in the H3N2 group and 5/9 (56%) subjects in the H1N1 group. For more details, see Supplementary Figure 1.
Antibody and T cell responses
All volunteers were screened to ensure they were seronegative for antibodies to the challenge virus. Antibody responses (HAI titer) were not detectable at 7 d when viruses were completely cleared from nasal lavage samples as indicated in Figure 1a, but antibody responses were detected at day 28 (Fig. 2a).
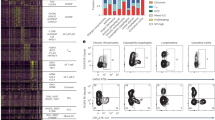
(a) Presence of influenza-specific antibody, as measured by hemagglutination inhibition assay (HAI). Error bars represent s.e.m. (b) Influenza-specific T lymphocyte responses measured from freshly isolated PBMC ex vivo from each volunteer by IFN-γ release after stimulation with the corresponding peptide pools spanning the entire challenge influenza proteome. Each bar represents the proportion of total T cell responses per million PBMCs in relation to the entire influenza proteome, and each color box represents the response to each protein. x axes denote subject number. HA, hemagglutinin; NA, neuraminidase; NP, nucleoprotein; M, matrix; NS, nonstructural proteins; PB1, PB2, PA, RNA polymerase complex. (c) Plot of relative proportion of CD4+ and CD8+ contribution to T cell responses to influenza peptides nucleoprotein and M at baseline for each cohort. Data from ex vivo stimulation and intracellular staining. (d) Activated and proliferating cells (CD38+Ki67+) detected ex vivo in H1N1-infected subjects by flow cytometry. Numbers represent activated, proliferating cells as a percentage of the total cell population. (e) Changes in the proportion of activated and proliferating (CD38+Ki67+ staining detected by flow cytometry) T cells in both CD4+ and CD8+ populations of freshly isolated PBMCs from volunteers infected with H1N1 virus. (f) Correlation between proportion of CD38+Ki67+ T cells (both CD4+ and CD8+) on day 7 of H1N1-infected subjects with their magnitude of ELISPOT response by Spearman rank correlation test.
Before viral challenge, we determined the nature of preexisting T cell memory. T cell responses to proteins expressed by the challenge virus were present in most volunteers in both studies before challenge despite the absence of detectable antibodies to these strains. The proportion of total T cell responses was below 1,000 spot-forming cells (SFCs) per million peripheral blood mononuclear cells (PBMCs) in all subjects studied at baseline (Fig. 2b). At baseline, in the H3N2 group, 11/14 (79%) infected subjects showed memory T cell responses recognizing one or more H3N2 proteins, with a median of two proteins recognized (range 1–5). The most immunodominant proteins were nucleoprotein (8/14, 57%) and matrix protein (7/14, 50%), as determined by the number of responding subjects and the magnitude of the interferon-γ (IFN-γ) response. These proteins are highly conserved across strains. In the H1N1 group, 7/9 (78%) infected subjects showed memory T cell responses that recognized one or more proteins at the baseline, with a median number of one protein recognized (range 1–3). The most immunodominant protein was matrix (6/9, 67%).
On day 7, the breadth and proportion of T cells responding to influenza peptides increased in the peripheral blood by an average of tenfold for both groups (Fig. 2b and Supplementary Fig. 2). In the H3N2 group, 14/14 (100%) infected subjects showed positive T cell responses, with a median of five proteins recognized (range 1–8). Preexisting T cell responses against each protein were expanded and new responses were detected that were absent at baseline. In the H1N1 group, 9/9 (100%) infected subjects were T cell response positive, responding to a median of five proteins (range 2–7). We found no significant changes from baseline in the day 7 T cell responses against known CD8+ epitopes of cytomegalovirus and Epstein-Barr virus in control wells, suggesting bystander activation was minimal (Supplementary Fig. 3).
On day 28, the total memory T cell response to challenge virus peptides returned to baseline levels (<1,000 SFCs per million PBMCs) in both groups (Supplementary Fig. 2). Immunodominant responses to proteins such as nucleoprotein and matrix persisted at a baseline level, whereas most newly generated responses against other proteins had vanished after the acute phase of infection. In the H3N2 group, 7/10 (70%) infected subjects that were sampled were T cell response positive, with the median number of proteins recognized reduced to 1 (range 1–2). In the H1N1 group, 8/9 infected subjects (89%) were T cell response positive, with the median number of proteins recognized reduced to 2 (range 2–4). However, 4/9 (44%) newly generated responses to hemagglutinin persisted at a lower level (average 60 SFCs per million PBMCs). Intracellular staining revealed that the responses to the immunodominant proteins (nucleoprotein and matrix) were predominantly mediated by CD4+ T cells in both groups (56% CD4 versus 44% CD8 for H3N2, and 72% CD4 versus 28% CD8 for H1N1) (Fig. 2c), consistent with a previous report12.
To understand the kinetics of T cell responses, we studied the functional status of both CD4+ and CD8+ T cells during the course of infection with H1N1 virus. Activated (CD38+) and proliferating (Ki67+) CD4+ and CD8+ T cells from freshly isolated PBMCs were undetectable before the challenge (Fig. 2d). Both markers were present in an expanded T cell population on day 7 before returning to baseline level on day 28 (Fig. 2e). The proportion of Ki67+CD38+ T cells correlated with the frequency of SFCs assayed by enzyme-linked immunospot (ELISPOT) on day 7 (Fig. 2f, r = 0.9, P = 0.002, Spearman coefficient).
Preexisting T cell responses, viral shedding and symptom scores
We studied the relationship between preexisting T cells responding to total influenza and immunodominant (nucleoprotein and matrix) proteins and virus control13 and illness severity14. The frequency of total preexisting T cells strongly correlated inversely with illness duration in the H3N2 study (Fig. 3a, r = −0.5740, P = 0.0318, Spearman coefficient) and with total symptom scores in the H1N1 study (Fig. 3b, r = −0.7113, P = 0.0369, Spearman coefficient).
(a,b) Correlations between influenza-specific total T and CD4+ T cell responses to internal proteins and measures of influenza severity (viral shedding, symptom severity or illness duration) in volunteers infected with H3N2 (WS/67/05) (a) or H1N1 (BR/59/07) (b). Correlations between total symptom scores or length of illness duration and influenza-specific T cell responses. Responses from total influenza-specific T cells and CD4+ T cells specific to internal proteins, nucleoprotein and matrix protein of challenge virus are shown separately. All tests were run by Spearman rank correlation test.
We found that the relationship between T cell responses to nucleoprotein and matrix and less severe illness was most closely associated with the number of preexisting CD4+ T cells (Fig. 3a,b), but not with CD8+ T cell responses (Supplementary Table 2a and Supplementary Fig. 4). The frequency of preexisting CD4+ but not CD8+ T cells recognizing the internal proteins nucleoprotein and matrix was inversely associated with total symptom scores in both groups. Notably, virus shedding of H3N2 predominantly and inversely correlated with the level of preexisting CD4+ T cell responses to nucleoprotein and matrix (r = −0.6087, P = 0.0209, Spearman coefficient), but this was not the case for preexisting CD8+ T cells (r = −0.0127, P = 0.9657, Spearman coefficient) (Supplementary Table 2 and Supplementary Fig. 4a).
To determine the relationship between the acutely expanding T cell population and illness metrics, we analyzed the relationship between peak T cell responses on day 7, viral load and symptom severity. The magnitude of the developing acute T cell response correlated positively with viral shedding (r = 0.7670, P = 0.0003), illness duration (r = 0.6270, P = 0.0071) and total symptom scores (r = 0.5045, P = 0.0389) in the H3N2 cohort (Supplementary Fig. 5). These findings suggest that preexisting memory CD4+ T cells are the key CMI correlate to limited illness severity and that, once illness is established, acutely expanding CD4+ T cell populations track peak viral load and symptoms.
Preexisting T cells against nucleoprotein and matrix protein
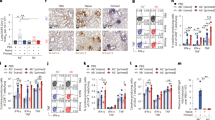
Preexisting T cell responses against internal proteins nucleoprotein and matrix measured by IFN-γ responses were largely CD4+ T cell mediated in both H1N1 and H3N2 study groups. In the H3N2 group, nine subjects had nucleoprotein and matrix responses at baseline, and 8/9 (89%) had their peptides identified at the single-peptide level (Table 1). For the matrix protein, 7/10 (70%) peptide responses were CD4+ T cell mediated, and for the nucleoprotein protein 8/11 (73%) peptide responses were CD4+ T cell mediated. In the H1N1 group, seven subjects' T cells responded to nucleoprotein and matrix at baseline, and 7/7 (100%) had their peptides identified at a single peptide level (Table 1). For the matrix protein, 5/5 (100%) peptides responses were CD4+ T cell mediated, and for the nucleoprotein protein it was 3/6 (50%) peptides. The antigen-specific T cell response on day 7 to nucleoprotein and matrix was mostly mediated by CD4+ T cells (Supplementary Table 3). Upregulation of CD107a expression on memory CD4+ T cells was observed following ex vivo stimulation with peptide pools to nucleoprotein or matrix protein (Fig. 4a).
(a) Expression of IFN-γ and CD107a in memory T cells of a representative H3N2-infected subject after stimulation with influenza protein peptide pools. PBMCs from baseline and day 7 samples were stimulated with different peptide pools (total influenza, NP, M) for 6 h, and the ex vivo response was measured by FACS staining. Both memory CD4+ and CD8+ responses in the same sample were measured. Staphylococcus enterotoxin B (SEB) was used as positive control. Numbers on plots represent percentage of total cells in each quadrant. (b) Killing function of CD4+ T cell lines from the same baseline sample upon recognition of autologous target cells pulsed with peptides was measured by chromium release assay. Perforin-dependent cytotoxicity was measured by sensitivity to concanamycin. E:T represents the effector to target cell ratio. 5A and 3A are individual subject identification numbers. M2 and NP14 are the influenza peptide identification numbers.
We further examined the cytotoxic function of these influenza-specific CD4+ cells15 by a 51Cr-release assay using peptide-pulsed autologous B cell lines and short-term T cell lines generated from baseline PBMCs. These CD4+ T cells killed pulsed autologous B cells in a peptide-specific manner (Fig. 4b). The killing was sensitive to concanamycin, suggesting cytotoxicity was dependent on the perforin-granzyme pathway. Similarly, the influenza-responsive CD8+ T cells also showed cytotoxic function in a concanamycin-sensitive manner (Supplementary Fig. 6a,b). The results were in agreement with a previous study in which most peptides derived from 2009 pandemic H1N1 virus were able to induce responses in seasonal influenza virus–specific T cells from pandemic H1N1–naive individuals16.
A role for cytotoxic CD4+ cells in limiting viral infection would imply the need for expression of major histocompatibility complex (MHC) class II on the respiratory epithelium, the target of influenza infection. To investigate this, we analyzed the constitutive expression of the MHC class II molecule HLA-DR in explanted lung tissue and on primary bronchial epithelial cells (PBECs) in culture and the effect of in vitro influenza infection on expression in PBECs. We found substantial constitutive expression of this molecule in both lung tissue and cultured PBECs, with a rise in HLA-DR expression after influenza infection of PBECs with X31 H3N2 compared to cells treated with ultraviolet light–inactivated virus (data in Supplementary Fig. 7a,b).
Heterosubtypic memory CD4+ T cell responses
We next tested whether these memory CD4+ T cells from H1N1 challenge subjects could cross-recognize equivalent peptides from pandemic H1N1 (A/Ca/07/2009) proteins on ex vivo ELISPOT. Of six selected subjects whose T cells responded to seasonal H1N1 (A/Brisbane/59/2007) peptides, T cells from five out of six also recognized peptide variants of pandemic H1N1 (A/Ca/07/2009), as determined by IFN-γ ELISPOT, despite several amino acid differences in the internal proteins (Fig. 5).
PBMCs from different donors were stimulated with decreasing concentrations of peptides, and the response to each peptide was measured by IFN-γ ELISPOT. Each plot represented the results from one donor to autologous peptide (closed circle) and heterologous peptide (open circles). Differences in peptide sequences between seasonal H1N1 (A/Brisbane/59/2007) and pandemic H1N1 (A/Ca/07/2009) are highlighted in bold, underlined text.
Discussion
We studied the nature of protective cell–mediated immunity against influenza infection in humans and identified a relationship between preexisting T cell immunity and illness severity in subjects seronegative for virus-specific antibody. We showed that the blood memory CD4+ T cell population that responded to peptides from core proteins of the challenge virus strains most closely correlated to limitation of illness severity, and that the CD4+ T cells presented cytotoxic characteristics.
Despite vaccination initiatives, influenza remains a major cause of mortality, morbidity and healthcare costs17,18,19. Currently, the majority of vaccines generate strain-specific serological protection using killed virus preparations20. The success of seasonal vaccination programs is dependent upon predictive modeling of strain circulation21 and the absence of viral coat protein mutation enabling immune evasion during an influenza season22,23. Furthermore, this strategy offers limited protection against pandemic strains, which may carry traits of high virulence and pathogenicity23,24,25.
Harnessing aspects of the immune response to offer protection against a broad range of circulating influenza strains and potentially an emergent pandemic strain would be a major advance. Previously, we have used a human challenge model similar to the one used here to establish a role for T cells in limiting influenza-associated illness8. Those subjects were not prescreened for protective antibodies to the challenge strain, although we considered the role of antibodies in the analysis. In subjects with an absence of strain-specific antibodies at the time of infection, cytotoxic T cell responses were associated with less virus shedding after challenge. However, CD8+ responses were not distinguished from CD4+ responses; the effector cells could have been either subset because the target cells used were autologous PBMCs activated with phytohemagglutinin (so, likely to express HLA class II). In our present study using more sophisticated methods, we have identified a relationship between influenza-specific CD4+ T lymphocytes and disease limitation.
In mice, cytotoxic CD8+ T lymphocytes have a crucial role in the response to influenza infection26,27. Although priming of naive CD8+ T cells is dependent on CD4+ T helper cell activity28 and CD4+ help is necessary for the maintenance of protective CD8+ memory T cell populations29, the role of CD4+ T cells as cytotoxic effectors is less certain. Recently, in a mouse model of herpes simplex infection, it was shown that CD4+ T cells have a vital role in the recruitment of cytotoxic CD8+ T cells into virally infected tissues30. Helper CD4+ T cells arrive at the site of infection before CD8+ T cells, secrete IFN-γ and induce expression of chemokines to attract CD8+ T cells. CD4+ T helper cells therefore may respond in an antigen-specific manner and orchestrate the effector T cell response. However, they may also act directly as antiviral cytotoxic T cells, as they stain positive for perforin and granzyme and produce IFN-γ31,32. This cytotoxic response at the site of infection may limit viral replication and disease severity. Cytotoxic CD4+ T cells have been found at low frequencies in the circulation of healthy donors but increase in number during viral infections32. They express a T helper type 1 phenotype, producing IFN-γ upon stimulation, but it is not known whether they require antigenic stimulation to maintain their numbers as do CD8+ T cells33.
Data from studies of natural influenza infection in humans and the role of CD4+ T cells in protective heterosubtypic immunity is scarce. A potential protective role for CD4+ T cells has been suggested by in vitro studies demonstrating reactivity against previously unencountered strains, for example, against avian H5N1 by CD4+ T cells primed with seasonal strains34,35,36,37. Further study of the role of CD4+ T cells in heterologous immunity is required.
Caution is required in comparing the results of a human study reliant on sampling T cells in the periphery to animal models where cellular immunity at the site of infection can be readily examined. The study of T cell responses in the lung during and after illness was beyond the technical feasibility of this study. Therefore, the role of CD8+ T cells and other cell types in the antiviral response cannot be discounted, and the relative contribution of different T cell types in the context of protective antibodies requires study. Demonstrating causal mechanisms in human studies is a complex process in comparison to mouse studies. However, future studies in which lung-resident or homing T cells are investigated directly by bronchial sampling techniques may offer insight into the relative contribution of CD4+ and CD8+ T cells in both orchestrating and effecting responses, and, subsequently, rational vaccine design may be optimized.
Understanding of the correlates of protection against influenza in humans remains limited. Current vaccines depend upon the use of killed viruses, which generate strain-specific antibody responses, or live attenuated viruses, which may offer some hope of heterosubtypic immunity38. Both technologies depend on the culture of live virus, which can limit the rate of vaccine development, as highlighted during the recent 2009 influenza pandemic. Technologies are available to produce vaccines using molecular techniques, but immunological readouts of efficacy are speculative, as true drivers of immune protection other than antibodies are uncertain39. We highlight a potential benefit of generating CD4+ T cell memory with recognition of peptides from core viral proteins that are tightly conserved across strains. Harnessing this response with appropriate technologies may provide an opportunity to produce inexpensive and effective vaccines with a broad spectrum of protection.
We used here a human challenge model of influenza infection. This has proven to be a safe and effective way to study responses to infection and new therapies in a manner that has previously been impossible in the field. The strains used for infection are closely related to circulating strains, although some loss of virulence is inevitable in their production during passage in eggs or cell lines. Consequently, confirmatory studies in the field are necessary but may be difficult to achieve. Furthermore, although we took care to ensure that all subjects were seronegative to the challenge strain, it remains possible that antibody activity against antigenically distinct strains from the challenge subtype may have contributed to the protective effects we observed. However, we did determine the heterologous humoral responses (to H3N2) in the H1N1 cohort at baseline (data not shown) and found that most subjects did show some antibody responses that were unrelated to illness metrics, unlike the CD4+ cellular response. This suggests that previous exposure to the heterologous strain had occurred but that heterologous adaptive immunity was related to CMI rather serological protection. Similarly, there was a possibility that subclinical infections from pandemic 2009 H1N1 could have had an impact on the A/Brisbane/59/07 H1N1 cohort study40. However, the likelihood of a substantial effect on our overall findings is minimal because the first cohort study was performed in 2008, well before the pandemic and the key mechanisms of protection were seen in both study cohorts. Also, the incidence of pandemic H1N1 infections described in field studies of this age group at the time were low, and the incidence of subclinical infections lower still40.
Further studies are required to determine whether future vaccines that generate CD4+ T cell responses to conserved core viral proteins may limit disease severity for a range of influenza viruses, including pandemic strains.
Methods
Study design.
We performed two separate prospective, randomized clinical studies of experimental human influenza A infections in a single site in the UK. The starting dates of the cohorts were 27 October 2008 for H3N2 and 21August 2009 for H1N1. Healthy, nonpregnant adults between the ages 18 and 45 were eligible for the enrollment. Exclusion criteria included history of respiratory illness, clinically significant co-morbidities or abnormal lung function.
We enrolled 17 volunteers with hemagglutination-inhibition titers less than 1:8 to influenza A/Wisconsin/67/05 into the H3N2 challenge study and 24 healthy adult volunteers with hemagglutination-inhibition titers less than 1:8 to influenza A/Brisbane/59/07 into the H1N1 challenge study.
We conducted the studies in compliance with Good Clinical Practice guidelines (CPMP/ICH/135/95) and the Declaration of Helsinki. The protocols were approved by East London and City and the Southampton and Southwest Hampshire Ethics Review Committees. Written informed consent was obtained from each participant with an ethics committee–approved form.
Study outline.
Screening assessments began within 45 d of the scheduled viral inoculation. Volunteers remained in isolation during the challenge phase of the study. We inoculated subjects intranasally with Good Manufacturing Practice grade virus on day 0 and collected nasal lavage daily during the quarantine period for virus isolation. Fresh whole blood for cellular assays was taken on day −2 and days 0, 7 and 28. An additional time point at day 3 was taken for the H1N1 study.
We recorded oral temperatures four times daily. Fever was defined as an oral temperature >37.7 °C. Symptom assessments were performed by the volunteers twice daily on a four-point scale (0–3 corresponding to absent to severe).
Virus titration by half-maximal tissue culture infectious dose assay.
We determined viral load in the nasal lavage samples by TCID50 assay as described by the World Health Organization manual of animal influenza41 by inoculating serial tenfold dilutions of virus-containing samples into 96-well plates seeded with Madin-Darby canine kidney cells incubated for 5–6 d at 37 °C. We assessed cytopathic effects in individual wells by light microscopy. Titers >1:5 were considered positive.
Hemagglutination-inhibition assay.
We determined hemagglutinin-specific antibody titers against H1N1 (A/Brisbane/59/2007) or H3N2 (A/Wisconsin/67/05) in the serum samples by hemagglutination-inhibition assay using chicken erythrocytes as described in the World Health Organization manual41.
Synthetic peptides.
We designed 18-amino-acid peptides overlapping by ten amino acid residues and spanning the full proteome of the H1N1 and H3N2 influenza A viruses using the Los Alamos National Library web-based software PeptGen (http://www.hiv.lanl.gov/content/sequence/PEPTGEN/peptgen.html) (purity >70%; PEPscreen; Sigma-Aldrich). In each influenza ELISPOT assay, we simultaneously tested all overlapping peptides using two-dimensional matrices with a total of 50 pools (first dimension: 25 pools; second dimension: 25 pools; up to 25 peptides per pool) so that each peptide was present in two different pools (see Supplementary Fig. 8 for ELISPOT layout) at a final concentration of 2 μg ml−1. We confirmed the putative peptides in the second ELISPOT assay with the same input cell number per well.
Ex vivo interferon-γ enzyme-linked immunospot.
We separated PBMCs from heparinized blood by density gradient centrifugation42. ELISPOT assays were performed using the human IFN-γ ELISPOT kit (Mabtech) according to the manufacturer's instructions. Negative and positive controls were run in parallel. We subtracted background T cell responses and presented values in SFCs per million PBMCs. To determine whether T cells were CD4+ or CD8+, in the second ELISPOT assay we conducted cell depletion by Dynal CD8 beads, as described in the manufacturer's instructions (Invitrogen) before the ELISPOT assay43,44. Supplementary Figure 9a–c illustrates the efficiency of the depletion methods used. Undepleted PBMCs served as positive controls. We generated short-term T cell lines to confirm influenza peptides and their CD4+ or CD8+ dependency by FACS staining assay, as described previously42.
FACS staining assay.
Activated (CD38+) and proliferating (Ki67+) cells in fresh PBMCs were stained with monoclonal antibodies against human Ki67 (conjugated to FITC, clone B56, BD Biosciences), HLA-DR (conjugated to phycoerythrin, clone TU36, BD), CD38 (conjugated to allophycocyanin, clone HB7, BD), CD4 (conjugated to Pacific blue, clone MT130, DakoCytomation) and CD8 (conjugated to phycoerythrin-Cy5, clone SK1, BD). Cytotoxicity was measured by expression of CD107a (clone H4A3, BD) and IFN-γ (clone XMG1.2, BD) in CD4+ and CD8+ cells and also ex vivo using frozen PBMCs, as described previously42. See Supplementary Figure 10 for gating strategy.
Chromium release assay.
We used a standard 51Cr release assay described previously12, with T cell lines generated from PBMCs as effector cells and autologous Epstein-Barr virus–transformed B cell lines as target cells. We inhibited perforin-mediated cytotoxicity by incubating the CD4+ T cells for 2 h with 100 nM concanamycin (Sigma).
Statistical analyses.
Statistical analysis used GraphPad Prism and SPSS. We presented the magnitude of T cell responses as SFCs per million PBMCs and the breadth as the number of proteins recognized by each subject. We made correlations between preexisting T cell readouts and measures of infection and illness using Spearman rank correlation analysis. Correlation analysis was based on data collected from all infected (culture positive and/or fourfold or greater rise in hemagglutination-inhibition antibody titer) individuals.
Additional methods.
Detailed methodology is described in the Supplementary Methods.
References
Holmes, E.C. Error thresholds and the constraints to RNA virus evolution. Trends Microbiol. 11, 543–546 (2003).
Both, G.W., Sleigh, M.J., Cox, N.J. & Kendal, A.P. Antigenic drift in influenza virus H3 hemagglutinin from 1968 to 1980: multiple evolutionary pathways and sequential amino acid changes at key antigenic sites. J. Virol. 48, 52–60 (1983).
Glezen, W.P. Emerging infections: pandemic influenza. Epidemiol. Rev. 18, 64–76 (1996).
Schweiger, B., Bruns, L. & Meixenberger, K. Reassortment between human A (H3N2) viruses is an important evolutionary mechanism. Vaccine 24, 6683–6690 (2006).
Stambas, J. et al. Killer T cells in influenza. Pharmacol. Ther. 120, 186–196 (2008).
Sun, J., Madan, R., Karp, C.L. & Braciale, T.J. Effector T cells control lung inflammation during acute influenza virus infection by producing IL-10. Nat. Med. 15, 277–284 (2009).
Kreijtz, J.H. et al. Primary influenza A virus infection induces cross-protective immunity against a lethal infection with a heterosubtypic virus strain in mice. Vaccine 25, 612–620 (2007).
McMichael, A.J., Gotch, F.M., Noble, G.R. & Beare, P.A. Cytotoxic T-cell immunity to influenza. N. Engl. J. Med. 309, 13–17 (1983).
Oxford, J.S., Gelder, C. & Lambkin, R. Would you volunteer to be quarantined and infected with influenza virus? Expert Rev. Anti Infect. Ther. 3, 1–2 (2005).
Ho, P.P.K., Young, A.L. & Truehaft, M. Plaque formation with influenza viruses in dog kidney cells. J. Gen. Virol. 33, 143–145 (1976).
Newton, D.W., Treanor, J.J. & Menegus, M.A. Clinical and laboratory diagnosis of influenza virus infections. Am. J. Manag. Care 6, S265–S275 (2000).
Lee, L.Y. et al. Memory T cells established by seasonal human influenza A infection cross-react with avian influenza A (H5N1) in healthy individuals. J. Clin. Invest. 118, 3478–3490 (2008).
Li, I.W. et al. The natural viral load profile of patients with pandemic 2009 influenza A(H1N1) and the effect of oseltamivir treatment. Chest 137, 759–768 (2010).
La Gruta, N.L., Kedzierska, K., Stambas, J. & Doherty, P.C. A question of self-preservation: immunopathology in influenza virus infection. Immunol. Cell Biol. 85, 85–92 (2007).
Casazza, J.P. et al. Acquisition of direct antiviral effector functions by CMV-specific CD4+ T lymphocytes with cellular maturation. J. Exp. Med. 203, 2865–2877 (2006).
Ge, X. et al. Assessment of seasonal influenza A virus–specific CD4 T-cell responses to 2009 pandemic H1N1 swine-origin influenza A virus. J. Virol. 84, 3312–3319 (2010).
Treanor, J. Influenza vaccine—outmaneuvering antigenic shift and drift. N. Engl. J. Med. 350, 218–220 (2004).
World Health Organization. Influenza (seasonal). 〈http://www.who.int/influenza/surveillance_monitoring/updates/latest_update_GIP_surveillance/en/index.html〉 (2009).
Monto, A.S. Epidemiology and virology of influenza illness. Am. J. Manag. Care 6, S255–S264 (2000).
Couch, R.B. An overview of serum antibody responses to influenza virus antigens. Dev. Biol. (Basel) 115, 25–30 (2003).
Aymard, M., Valette, M., Lina, B. & Thouvenot, D. Surveillance and impact of influenza in Europe. Groupe Regional d'Observation de la Grippe and European Influenza Surveillance Scheme. Vaccine 17, S30–S41 (1999).
Creanza, N., Schwarz, J.S. & Cohen, J.E. Intraseasonal dynamics and dominant sequences in H3N2 influenza. PLoS ONE 5, e8544 (2010).
Shinde, V. et al. Triple-reassortant swine influenza A (H1) in humans in the United States, 2005–2009. N. Engl. J. Med. 360, 2616–2625 (2009).
Belshe, R.B. The origins of pandemic influenza—lessons from the 1918 virus. N. Engl. J. Med. 353, 2209–2211 (2005).
Kash, J.C. et al. Genomic analysis of increased host immune and cell death responses induced by 1918 influenza virus. Nature 443, 578–581 (2006).
Topham, D.J., Tripp, R.A. & Doherty, P.C. CD8+ T cells clear influenza virus by perforin or Fas-dependent processes. J. Immunol. 159, 5197–5200 (1997).
Yang, O.O. et al. Efficient lysis of human immunodeficiency virus type 1-infected cells by cytotoxic T lymphocytes. J. Virol. 70, 5799–5806 (1996).
Bevan, M.J. Helping the CD8+ T-cell response. Nat. Rev. Immunol. 4, 595–602 (2004).
Kalams, S.A. & Walker, B.D. The critical need for CD4 help in maintaining effective cytotoxic T lymphocyte responses. J. Exp. Med. 188, 199–2204 (1998).
Nakanishi, Y., Lu, B., Gerard, C. & Iwasaki, A. CD8+ T lymphocyte mobilization to virus-infected tissue requires CD4+ T-cell help. Nature 462, 510–513 (2009).
van de Berg, P.J., van Leeuwen, E.M., ten Berge, I.J. & van Lier, R. Cytotoxic human CD4+ cells. Curr. Opin. Immunol. 20, 339–343 (2008).
Appay, V. et al. Characterization of CD4+ CTLs ex vivo. J. Immunol. 168, 5954–5958 (2002).
Shin, H., Blackburn, S.D., Blattman, J.N. & Wherry, E.J. Viral antigen and extensive division maintain virus-specific CD8 T cells during chronic infection. J. Exp. Med. 204, 941–949 (2007).
McKinstry, K.K., Strutt, T.M. & Swain, S.L. Hallmarks of CD4 T cell immunity against influenza. J. Intern. Med. 269, 507–518 (2011).
Richards, K.A., Topham, D., Chaves, F.A. & Sant, A.J. Cutting edge: CD4 T cells generated from encounter with seasonal influenza viruses and vaccines have broad protein specificity and can directly recognize naturally generated epitopes derived from the live pandemic H1N1 virus. J. Immunol. 185, 4998–5002 (2010).
Lee, L.Y. et al. Memory T cells established by seasonal human influenza A infection cross-react with avian influenza A (H5N1) in healthy individuals. J. Clin. Invest. 118, 3478–3490 (2008).
Roti, M. et al. Healthy human subjects have CD4+ T cells directed against H5N1 influenza virus. J. Immunol. 180, 1758–1768 (2008).
Belshe, R.B. & Mendelman, P.M. Safety and efficacy of live attenuated, cold-adapted, influenza vaccine-trivalent. Immunol. Allergy Clin. North Am. 23, 745–767 (2003).
Rimmelzwaan, G.F. & McElhaney, J.E. Correlates of protection: novel generations of influenza vaccines. Vaccine 26, D41–D44 (2008).
Riley, S. et al. Epidemiological characteristics of 2009 (H1N1) pandemic influenza based on paired sera from a longitudinal community cohort study. PLoS Med. 8, e1000442 (2011).
World Health Organization Global Surveillance Network. Manual for the laboratory diagnosis and virological surveillance of influenza. http://whqlibdoc.who.int/publications/2011/9789241548090_eng.pdf (2011).
Li, C.K. et al. T cell responses to whole SARS coronavirus in humans. J. Immunol. 181, 5490–5500 (2008).
Tobery, T.W. & Caulfield, M.J. Identification of T-cell epitopes using ELISPOT and peptide pool arrays. Methods Mol. Med. 94, 121–132 (2004).
Kaufhold, R.M. et al. Memory T cell response to rotavirus detected with a gamma interferon enzyme-linked immunospot assay. J. Virol. 79, 5684–5694 (2005).
Acknowledgements
We thank M. Roederer, P. Chattopadhyay and J. Yu (US National Institutes of Health) for providing the antibody conjugates used for flow cytometry . We also thank G. Balaratnam and the Retroscreen Clinical team for their expert conduct of the clinical study. In addition, we thank N. Goonetilleke for discussion of the statistics, H. Hong and L. Wang for assistance in blood separation and ELISPOT assay, and J. Ward of the Southampton Histochemistry Research Unit for assistance with immunohistochemistry. This study was partially funded by the UK Medical Research Council.
Author information
Authors and Affiliations
Contributions
T.M.W. designed the clinical study and supervised its conduct, performed the analysis and wrote the manuscript. C.K.F.L., D.C.D., A.J.M. and X.-N.X. designed the experiments, performed the analysis and wrote the manuscript. C.S.C.C., A.K.Y.H., M.P., J.C.L., B.N., K.J.S. and T.D. performed the laboratory experiments and assays. R.L.-W., J.O. and A.G. designed and supervised the clinical studies.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–10, Supplementary Tables 1–3 and Supplementary Methods (PDF 1319 kb)
Rights and permissions
About this article
Cite this article
Wilkinson, T., Li, C., Chui, C. et al. Preexisting influenza-specific CD4+ T cells correlate with disease protection against influenza challenge in humans. Nat Med 18, 274–280 (2012). https://doi.org/10.1038/nm.2612
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nm.2612
This article is cited by
-
Therapeutic mitigation of measles-like immune amnesia and exacerbated disease after prior respiratory virus infections in ferrets
Nature Communications (2024)
-
Analysis of clinical characteristics and risk factors for death due to severe influenza in children
European Journal of Clinical Microbiology & Infectious Diseases (2024)
-
A polyvalent virosomal influenza vaccine induces broad cellular and humoral immunity in pigs
Virology Journal (2023)
-
Human circulating and tissue-resident memory CD8+ T cells
Nature Immunology (2023)
-
Respiratory mucosal immune memory to SARS-CoV-2 after infection and vaccination
Nature Communications (2023)