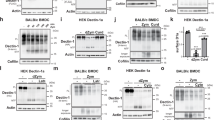
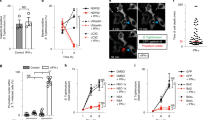
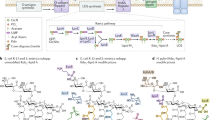
Abstract
Mammalian peptidoglycan recognition proteins (PGRPs), similar to antimicrobial lectins, bind the bacterial cell wall and kill bacteria through an unknown mechanism. We show that PGRPs enter the Gram-positive cell wall at the site of daughter cell separation during cell division. In Bacillus subtilis, PGRPs activate the CssR-CssS two-component system that detects and disposes of misfolded proteins that are usually exported out of bacterial cells. This activation results in membrane depolarization, cessation of intracellular peptidoglycan, protein, RNA and DNA synthesis, and production of hydroxyl radicals, which are responsible for bacterial death. PGRPs also bind the outer membrane of Escherichia coli and activate the functionally homologous CpxA-CpxR two-component system, which kills the bacteria. We exclude other potential bactericidal mechanisms, including inhibition of extracellular peptidoglycan synthesis, hydrolysis of peptidoglycan and membrane permeabilization. Thus, we reveal a previously unknown mechanism by which innate immunity proteins that bind the cell wall or outer membrane exploit the bacterial stress defense response to kill bacteria.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Cash, H.L., Whitham, C.V., Behrendt, C.L. & Hooper, L.V. Symbiotic bacteria direct expression of an intestinal bactericidal lectin. Science 313, 1126–1130 (2006).
Stowell, S.R. et al. Innate immune lectins kill bacteria expressing blood group antigen. Nat. Med. 16, 295–301 (2010).
Dziarski, R. & Gupta, D. The peptidoglycan recognition proteins (PGRPs). Genome Biol. 7, 232 (2006).
Royet, J. & Dziarski, R. Peptidoglycan recognition proteins: pleiotropic sensors and effectors of antimicrobial defenses. Nat. Rev. Microbiol. 5, 264–277 (2007).
Dziarski, R. & Gupta, D. Mammalian PGRPs: novel antibacterial proteins. Cell. Microbiol. 8, 1059–1069 (2006).
Kang, D., Liu, G., Lundstrom, A., Gelius, E. & Steiner, H. A peptidoglycan recognition protein in innate immunity conserved from insects to humans. Proc. Natl. Acad. Sci. USA 95, 10078–10082 (1998).
Liu, C., Xu, Z., Gupta, D. & Dziarski, R. Peptidoglycan recognition proteins: a novel family of four human innate immunity pattern recognition molecules. J. Biol. Chem. 276, 34686–34694 (2001).
Tydell, C.C., Yount, N., Tran, D., Yuan, J. & Selsted, M. Isolation, characterization and antimicrobial properties of bovine oligosaccharide-binding protein. J. Biol. Chem. 277, 19658–19664 (2002).
Dziarski, R., Platt, K.A., Gelius, E., Steiner, H. & Gupta, D. Defect in neutrophil killing and increased susceptibility to infection with non-pathogenic Gram-positive bacteria in peptidoglycan recognition protein-S (PGRP-S)-deficient mice. Blood 102, 689–697 (2003).
Lu, X. et al. Peptidoglycan recognition proteins are a new class of human bactericidal proteins. J. Biol. Chem. 281, 5895–5907 (2006).
Tydell, C.C., Yuan, J., Tran, P. & Selsted, M.E. Bovine peptidoglycan recognition protein-S: antimicrobial activity, localization, secretion and binding properties. J. Immunol. 176, 1154–1162 (2006).
Wang, M. et al. Human peptidoglycan recognition proteins require zinc to kill both Gram-positive and Gram-negative bacteria and are synergistic with antibacterial peptides. J. Immunol. 178, 3116–3125 (2007).
Gelius, E., Persson, C., Karlsson, J. & Steiner, H. A mammalian peptidoglycan recognition protein with N-acetylmuramoyl-L-alanine amidase activity. Biochem. Biophys. Res. Commun. 306, 988–994 (2003).
Wang, Z.-M. et al. Human peptidoglycan recognition protein-L is an N-acetylmuramoyl-L-alanine amidase. J. Biol. Chem. 278, 49044–49052 (2003).
Cho, S. et al. Structural insights into the bactericidal mechanism of human peptidoglycan recognition proteins. Proc. Natl. Acad. Sci. USA 104, 8761–8766 (2007).
Hancock, R.E.W. & Sahl, H.-G. Antimicrobial and host-defense peptides as new anti-infective therapeutic strategies. Nat. Biotechnol. 24, 1551–1557 (2006).
Peschel, A. & Sahl, H.-G. The co-evolution of host cationic antimicrobial peptides and microbial resistance. Nat. Rev. Microbiol. 4, 529–536 (2006).
Fukushima, T. et al. A new D,L-endopeptidase gene product, YojL (renamed CwlS), plays a role in cell separation with LytE and LytF in Bacillus subtilis. J. Bacteriol. 188, 5541–5550 (2006).
Yamamoto, H., Kurosawa, S. & Sekiguchi, J. Localization of the vegetative cell wall hydrolases LytC, LytE and LytF on the Bacillus subtilis cell surface and stability of these enzymes to cell wall-bound or extracellular proteases. J. Bacteriol. 185, 6666–6677 (2003).
Blackman, S.A., Smith, T.J. & Foster, S.J. The role of autolysins during vegetative growth of Bacillus subtilis 168. Microbiology 144, 73–82 (1998).
Kohanski, M.A., Dwyer, D.J., Hayete, B., Lawrence, C.A. & Collins, J.J. A common mechanism of cellular death induced by bactericidal antibiotics. Cell 130, 797–810 (2007).
Hyyryläinen, H.L. et al. A novel two-component regulatory system in Bacillus subtilis for the survival of severe secretion stress. Mol. Microbiol. 41, 1159–1172 (2001).
Kohanski, M.A., Dwyer, D.J., Wierzbowski, J., Cottarel, G. & Collins, J.J. Mistranslation of membrane proteins and two-component system activation trigger antibiotic-mediated cell death. Cell 135, 679–690 (2008).
DiGiuseppe, P.A. & Silhavy, T.J. Signal detection and target gene induction by the CpxRA two-component system. J. Bacteriol. 185, 2432–2440 (2003).
Danese, P.N., Snyder, W.B., Cosma, C.L., Davis, L.J. & Silhavy, T.J. The Cpx two-component signal transduction pathway of Escherichia coli regulates transcription of the gene specifying the stress-inducible periplasmic protease, DegP. Genes Dev. 9, 387–398 (1995).
Nash, J.A., Ballard, T.N., Weaver, T.E. & Akinbi, H.T. The peptidoglycan-degrading property of lysozyme is not required for bactericidal activity in vivo. J. Immunol. 177, 519–526 (2006).
Darmon, E. et al. A novel class of heat and secretion stress-responsive genes is controlled by the autoregulated CssRS two-component system of Bacillus subtilis. J. Bacteriol. 184, 5661–5671 (2002).
Westers, H. et al. The CssRS two-component regulatory system controls a general secretion stress response in Bacillus subtilis. FEBS J. 273, 3816–3827 (2006).
Lim, J.H. et al. Structural basis for preferential recognition of diaminopimelic acid–type peptidoglycan by a subset of peptidoglycan recognition proteins. J. Biol. Chem. 281, 8286–8295 (2006).
Touhami, A., Jericho, M.H. & Beveridge, T.J. Atomic force microscopy of cell growth and division in Staphylococcus aureus. J. Bacteriol. 186, 3286–3295 (2004).
Yamada, S. et al. An autolysin ring associated with cell separation of Staphylococcus aureus. J. Bacteriol. 178, 1565–1571 (1996).
Chang, C.I., Chelliah, Y., Borek, D., Mengin-Lecreulx, D. & Deisenhofer, J. Structure of tracheal cytotoxin in complex with a heterodimeric pattern-recognition receptor. Science 311, 1761–1764 (2006).
Kim, M.-S., Byun, M. & Oh, B.-H. Crystal structure of peptidoglycan recognition protein LB from Drosophila melanogaster. Nat. Immunol. 4, 787–793 (2003).
Liu, C., Gelius, E., Liu, G., Steiner, H. & Dziarski, R. Mammalian peptidoglycan recognition protein binds peptidoglycan with high affinity, is expressed in neutrophils and inhibits bacterial growth. J. Biol. Chem. 275, 24490–24499 (2000).
Dwyer, D.J., Kohanski, M.A., Hayete, B. & Collins, J.J. Gyrase inhibitors induce an oxidative damage cellular death pathway in Escherichia coli. Mol. Syst. Biol. 3, 91 (2007).
Meijer, W.J. et al. The endogenous Bacillus subtilis (natto) plasmids pTA1015 and pTA1040 contain signal peptidase-encoding genes: identification of a new structural module on cryptic plasmids. Mol. Microbiol. 17, 621–631 (1995).
Acknowledgements
We are grateful to J.M. van Dijl, O.P. Kuipers and V.P. Kontinen and their associates J. Zweers and S. Holsappel (University of Groningen and National Institute of Health and Welfare Finland), J. Sekiguchi (Shinshu University) and S.J. Foster (University of Sheffield) for providing B. subtilis mutants; to J.J. Collins and M.A. Kohanski (Boston University) for providing E. coli mutants; to J.M. van Dijl and his associates T. Kouwen and M. Sibbald (University of Groningen) for the pGDL48 plasmid and its sequence; to M. Wang for analyzing samples by mass spectrometry; and to Huvepharma for providing moenomycin. This work was supported by the US Public Health Service grants from the US National Institutes of Health AI073290 and AI028797 to R.D. and D.G. and GM061761 to G.-J.B.
Author information
Authors and Affiliations
Contributions
D.R.K., M.W., D.G. and R.D. designed the experiments, D.R.K., M.W. and R.D. performed the experiments, L.-H.L. and D.R.K. obtained and purified the proteins, G.-J.B. synthesized muramyl peptides, and R.D. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–8, Supplementary Results, Supplementary Discussion, Supplementary Methods and Supplementary Table 1 (PDF 1852 kb)
Rights and permissions
About this article
Cite this article
Kashyap, D., Wang, M., Liu, LH. et al. Peptidoglycan recognition proteins kill bacteria by activating protein-sensing two-component systems. Nat Med 17, 676–683 (2011). https://doi.org/10.1038/nm.2357
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nm.2357
This article is cited by
-
Triggering receptor expressed on myeloid cells-1 in sepsis, and current insights into clinical studies
Critical Care (2024)
-
Pattern Recognition Beyond the Surface: Soluble Pattern Recognition and Their Role in Periodontitis
Current Oral Health Reports (2022)
-
Respiratory chain components are required for peptidoglycan recognition protein-induced thiol depletion and killing in Bacillus subtilis and Escherichia coli
Scientific Reports (2021)
-
Formate dehydrogenase, ubiquinone, and cytochrome bd-I are required for peptidoglycan recognition protein-induced oxidative stress and killing in Escherichia coli
Scientific Reports (2020)
-
A novel electrochemical biosensor based on peptidoglycan and platinum-nickel-copper nano-cube for rapid detection of Gram-positive bacteria
Microchimica Acta (2020)