Abstract
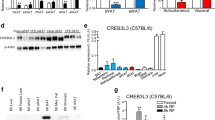
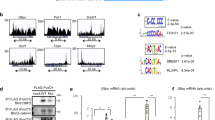
Here we report that the transcription factor cyclic AMP–responsive element–binding protein H (CREB-H, encoded by CREB3L3) is required for the maintenance of normal plasma triglyceride concentrations. CREB-H–deficient mice showed hypertriglyceridemia secondary to inefficient triglyceride clearance catalyzed by lipoprotein lipase (Lpl), partly due to defective expression of the Lpl coactivators Apoc2, Apoa4 and Apoa5 (encoding apolipoproteins C2, A4 and A5, respectively) and concurrent augmentation of the Lpl inhibitor Apoc3. We identified multiple nonsynonymous mutations in CREB3L3 that produced hypomorphic or nonfunctional CREB-H protein in humans with extreme hypertriglyceridemia, implying a crucial role for CREB-H in human triglyceride metabolism.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
Accession codes
References
Luebke-Wheeler, J. et al. Hepatology 48, 1242–1250 (2008).
Omori, Y. et al. Nucleic Acids Res. 29, 2154–2162 (2001).
Zhang, K. et al. Cell 124, 587–599 (2006).
Bailey, D., Barreca, C. & O'Hare, P. Traffic 8, 1796–1814 (2007).
Danno, H. et al. Biochem. Biophys. Res. Commun. 391, 1222–1227 (2010).
Lee, M.W. et al. Cell Metab. 11, 331–339 (2010).
Merkel, M., Eckel, R.H. & Goldberg, I.J. J. Lipid Res. 43, 1997–2006 (2002).
Hegele, R.A. Nat. Rev. Genet. 10, 109–121 (2009).
Kliewer, S.A. & Mangelsdorf, D.J. Am. J. Clin. Nutr. 91, 254S–257S (2010).
Guillou, H., Zadravec, D., Martin, P.G. & Jacobsson, A. Prog. Lipid Res. 49, 186–199 (2010).
Gong, J., Sun, Z. & Li, P. Curr. Opin. Lipidol. 20, 121–126 (2009).
Yang, X. et al. Cell Metab. 11, 194–205 (2010).
Goldberg, I.J., Scheraldi, C.A., Yacoub, L.K., Saxena, U. & Bisgaier, C.L. J. Biol. Chem. 265, 4266–4272 (1990).
Jong, M.C., Hofker, M.H. & Havekes, L.M. Arterioscler. Thromb. Vasc. Biol. 19, 472–484 (1999).
Bodmer, W. & Bonilla, C. Nat. Genet. 40, 695–701 (2008).
Johansen, C.T., Kathiresan, S. & Hegele, R.A. J. Lipid Res. 52, 189–206 (2011).
Johansen, C.T. et al. Nat. Genet. 42, 684–687 (2010).
1000 Genomes Project Consortium. Nature 467, 1061–1073 (2010).
Horton, J.D., Goldstein, J.L. & Brown, M.S. J. Clin. Invest. 109, 1125–1131 (2002).
Duval, C., Muller, M. & Kersten, S. Biochim. Biophys. Acta 1771, 961–971 (2007).
Acknowledgements
We thank B. Zhai and S. Gygi for mass spectrometry analysis, K. Sigrist for assistance with the development of transgenic mice, R. Hassell for assistance with genomic DNA sequencing, B. Monia (Isis Innovation) for Apoc3-specific antibody and D. Cohen for invaluable suggestions and comments on the manuscript. The research was supported by the Harvard University Accelerator Fund (L.H.G.), a Howard Hughes Medical Institute (HHMI) fellowship (P.G.), the Harvard Digestive Disease Center, US National Institutes of Health grant P30 DK34845 (A.-H.L.), US National Institutes of Health grant DK082448 (L.H.G.) and grants from the American Heart Association (A.-H.L. and J.H.L.), Canadian Institutes for Health Research and Genome Canada through the Ontario Genomics Institute.
Author information
Authors and Affiliations
Contributions
A.-H.L. designed the experiments. J.H.L. performed in vivo experiments in Creb3l3−/− mice and mutational analysis of CREB-H protein. A.-H.L. and P.G. generated and characterized CREB-H(N)-transgenic mice. J.D.B. performed post-heparin LPL assays. J.W. and C.T.J. performed sequencing experiments. S.A.D. provided Creb3l3−/− mice. A.-H.L., L.H.G., R.A.H. and J.P. analyzed the data. A.-H.L., L.H.G. and R.A.H. wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
L.H.G. holds equity in and is on the corporate board of directors of Bristol-Myers Squibb.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–5, Supplementary Tables 1 and 2 and Supplementary Methods (PDF 1842 kb)
Rights and permissions
About this article
Cite this article
Lee, J., Giannikopoulos, P., Duncan, S. et al. The transcription factor cyclic AMP–responsive element–binding protein H regulates triglyceride metabolism. Nat Med 17, 812–815 (2011). https://doi.org/10.1038/nm.2347
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nm.2347
This article is cited by
-
CREB-H is a stress-regulator of hepcidin gene expression during early postnatal development
Journal of Molecular Medicine (2023)
-
Adipose expression of CREB3L3 modulates body weight during obesity
Scientific Reports (2021)
-
Mechanisms, regulation and functions of the unfolded protein response
Nature Reviews Molecular Cell Biology (2020)
-
Hepatocyte CREBH deficiency aggravates inflammatory liver injury following chemokine-dependent neutrophil infiltration through upregulation of NF-κB p65 in mice
Archives of Toxicology (2020)
-
The Role and Potential Therapeutic Implications of the Fibroblast Growth Factors in Energy Balance and Type 2 Diabetes
Current Diabetes Reports (2017)