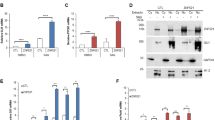
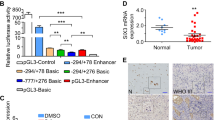
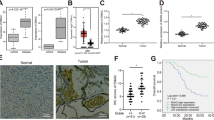
Abstract
Aberrant activation of the Hedgehog (Hh) pathway can drive tumorigenesis1. To investigate the mechanism by which glioma-associated oncogene family zinc finger-1 (GLI1), a crucial effector of Hh signaling2, regulates Hh pathway activation, we searched for GLI1-interacting proteins. We report that the chromatin remodeling protein SNF5 (encoded by SMARCB1, hereafter called SNF5), which is inactivated in human malignant rhabdoid tumors (MRTs), interacts with GLI1. We show that Snf5 localizes to Gli1-regulated promoters and that loss of Snf5 leads to activation of the Hh-Gli pathway. Conversely, re-expression of SNF5 in MRT cells represses GLI1. Consistent with this, we show the presence of a Hh-Gli–activated gene expression profile in primary MRTs and show that GLI1 drives the growth of SNF5-deficient MRT cells in vitro and in vivo. Therefore, our studies reveal that SNF5 is a key mediator of Hh signaling and that aberrant activation of GLI1 is a previously undescribed targetable mechanism contributing to the growth of MRT cells.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Jiang, J. & Hui, C.C. Hedgehog signaling in development and cancer. Dev. Cell 15, 801–812 (2008).
Ruiz i Altaba, A., Mas, C. & Stecca, B. The Gli code: an information nexus regulating cell fate, stemness and cancer. Trends Cell Biol. 17, 438–447 (2007).
Gailani, M.R. et al. The role of the human homologue of Drosophila patched in sporadic basal cell carcinomas. Nat. Genet. 14, 78–81 (1996).
Raffel, C. et al. Sporadic medulloblastomas contain PTCH mutations. Cancer Res. 57, 842–845 (1997).
Kinzler, K.W. et al. Identification of an amplified, highly expressed gene in a human glioma. Science 236, 70–73 (1987).
Northcott, P.A. et al. Multiple recurrent genetic events converge on control of histone lysine methylation in medulloblastoma. Nat. Genet. 41, 465–472 (2009).
Kogerman, P. et al. Mammalian suppressor-of-fused modulates nuclear-cytoplasmic shuttling of Gli-1. Nat. Cell Biol. 1, 312–319 (1999).
Roberts, C.W. & Orkin, S.H. The SWI/SNF complex–chromatin and cancer. Nat. Rev. Cancer 4, 133–142 (2004).
Weissman, B. & Knudsen, K.E. Hijacking the chromatin remodeling machinery: impact of SWI/SNF perturbations in cancer. Cancer Res. 69, 8223–8230 (2009).
Wu, J.I., Lessard, J. & Crabtree, G.R. Understanding the words of chromatin regulation. Cell 136, 200–206 (2009).
Goodrich, L.V., Johnson, R.L., Milenkovic, L., McMahon, J.A. & Scott, M.P. Conservation of the hedgehog/patched signaling pathway from flies to mice: induction of a mouse patched gene by Hedgehog. Genes Dev. 10, 301–312 (1996).
Lee, J., Platt, K.A., Censullo, P. & Ruiz i Altaba, A. Gli1 is a target of Sonic hedgehog that induces ventral neural tube development. Development 124, 2537–2552 (1997).
Isakoff, M.S. et al. Inactivation of the Snf5 tumor suppressor stimulates cell cycle progression and cooperates with p53 loss in oncogenic transformation. Proc. Natl. Acad. Sci. USA 102, 17745–17750 (2005).
Logan, M. et al. Expression of Cre recombinase in the developing mouse limb bud driven by a Prxl enhancer. Genesis 33, 77–80 (2002).
Martin, J.F. & Olson, E.N. Identification of a prx1 limb enhancer. Genesis 26, 225–229 (2000).
Theil, T., Kaesler, S., Grotewold, L., Bose, J. & Ruther, U. Gli genes and limb development. Cell Tissue Res. 296, 75–83 (1999).
Wang, X. et al. Oncogenesis caused by loss of the SNF5 tumor suppressor is dependent on activity of BRG1, the ATPase of the SWI/SNF chromatin remodeling complex. Cancer Res. 69, 8094–8101 (2009).
Biegel, J.A. et al. Germ-line and acquired mutations of INI1 in atypical teratoid and rhabdoid tumors. Cancer Res. 59, 74–79 (1999).
Sévenet, N. et al. Constitutional mutations of the hSNF5/INI1 gene predispose to a variety of cancers. Am. J. Hum. Genet. 65, 1342–1348 (1999).
Versteege, I. et al. Truncating mutations of hSNF5/INI1 in aggressive paediatric cancer. Nature 394, 203–206 (1998).
McKenna, E.S. et al. Loss of the epigenetic tumor suppressor SNF5 leads to cancer without genomic instability. Mol. Cell. Biol. 28, 6223–6233 (2008).
McKenna, E.S. & Roberts, C.W. Epigenetics and cancer without genomic instability. Cell Cycle 8, 23–26 (2009).
Pomeroy, S.L. et al. Prediction of central nervous system embryonal tumour outcome based on gene expression. Nature 415, 436–442 (2002).
Wisniewski, D. et al. Characterization of potent inhibitors of the Bcr-Abl and the c-kit receptor tyrosine kinases. Cancer Res. 62, 4244–4255 (2002).
Hyman, J.M. et al. Small-molecule inhibitors reveal multiple strategies for Hedgehog pathway blockade. Proc. Natl. Acad. Sci. USA 106, 14132–14137 (2009).
Pan, S. et al. Discovery of NVP-LDE225, a potent and selective smoothened antagonist. ACS Med. Chem. Lett. 1, 130–134 (2010).
Chai, J., Charboneau, A.L., Betz, B.L. & Weissman, B.E. Loss of the hSNF5 gene concomitantly inactivates p21CIP/WAF1 and p16INK4a activity associated with replicative senescence in A204 rhabdoid tumor cells. Cancer Res. 65, 10192–10198 (2005).
Lee, D. et al. SWI/SNF complex interacts with tumor suppressor p53 and is necessary for the activation of p53-mediated transcription. J. Biol. Chem. 277, 22330–22337 (2002).
Nagl, N.G. Jr., Zweitzig, D.R., Thimmapaya, B., Beck, G.R. Jr. & Moran, E. The c-myc gene is a direct target of mammalian SWI/SNF-related complexes during differentiation-associated cell cycle arrest. Cancer Res. 66, 1289–1293 (2006).
Tsikitis, M., Zhang, Z., Edelman, W., Zagzag, D. & Kalpana, G.V. Genetic ablation of cyclin D1 abrogates genesis of rhabdoid tumors resulting from Ini1 loss. Proc. Natl. Acad. Sci. USA 102, 12129–12134 (2005).
Rudin, C.M. et al. Treatment of medulloblastoma with hedgehog pathway inhibitor GDC-0449. N. Engl. J. Med. 361, 1173–1178 (2009).
Bouwmeester, T. et al. A physical and functional map of the human TNF-α/NF-κB signal transduction pathway. Nat. Cell Biol. 6, 97–105 (2004).
Frank-Kamenetsky, M. et al. Small-molecule modulators of Hedgehog signaling: identification and characterization of Smoothened agonists and antagonists. J. Biol. 1, 10 (2002).
Turner, F.B., Cheung, W.L. & Cheung, P. Chromatin immunoprecipitation assay for mammalian tissues. Methods Mol. Biol. 325, 261–272 (2006).
Roberts, C.W., Leroux, M.M., Fleming, M.D. & Orkin, S.H. Highly penetrant, rapid tumorigenesis through conditional inversion of the tumor suppressor gene Snf5. Cancer Cell 2, 415–425 (2002).
Riddle, R.D., Johnson, R.L., Laufer, E. & Tabin, C. Sonic hedgehog mediates the polarizing activity of the ZPA. Cell 75, 1401–1416 (1993).
Irizarry, R.A. et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4, 249–264 (2003).
Barbie, D.A. et al. Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature 462, 108–112 (2009).
Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 102, 15545–15550 (2005).
Acknowledgements
We thank X. Wang, A. Bouret, J. DaSilva and A. Carlson for their assistance and S. S. Kadam for critical advice. The Gli1 probe was generously provided by C. Tabin (Harvard University Medical School).
Author information
Authors and Affiliations
Contributions
Z.J. initiated the studies, conducted most of the experiments, analyzed data and wrote the manuscript. E.L.M.-B. conducted the Gli1 in situ experiments. C.G.S. generated Snf5-inactivated MEFs. E.S.M. contributed to Snf5 re-expression studies. B.W. assisted with ChIP studies. D.C. and J.M. conducted in vivo experiments. J.K. performed statistical analysis from interaction screen. P.T., Y.-J.C., J.P.M. and S.L.P. contributed to the gene expression analysis. M.T. and P.J.P. performed nucleosome repositioning assays. H.R., T.B., S.J.L. and J.F.K. contributed to the TAP-protein interaction screen. P.T.L.N. contributed to GLI shRNA studies. K.H. generated vectors for GLI1 deletions. S.B. contributed to the writing of the manuscript. M.W., W.R.S., C.W.M.R. and M.D. supervised the studies, assisted in data analysis and contributed to the writing of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
Z.J., D.C., J.P.M., K.H., S.B., J.K., H.R., T.B., S.L., J.F.K., M.W., and W.R.S. are employees of the Novartis Institutes for BioMedical Research (NIBR).
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–11 and Supplementary Methods (PDF 1013 kb)
Rights and permissions
About this article
Cite this article
Jagani, Z., Mora-Blanco, E., Sansam, C. et al. Loss of the tumor suppressor Snf5 leads to aberrant activation of the Hedgehog-Gli pathway. Nat Med 16, 1429–1433 (2010). https://doi.org/10.1038/nm.2251
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nm.2251
This article is cited by
-
Hedgehog pathway in sarcoma: from preclinical mechanism to clinical application
Journal of Cancer Research and Clinical Oncology (2023)
-
Reciprocal FGF19-GLI2 signaling induces epithelial-to-mesenchymal transition to promote lung squamous cell carcinoma metastasis
Cellular Oncology (2023)
-
ATRT–SHH comprises three molecular subgroups with characteristic clinical and histopathological features and prognostic significance
Acta Neuropathologica (2022)
-
SNF5 as a prognostic factor in skull base chordoma
Journal of Neuro-Oncology (2018)