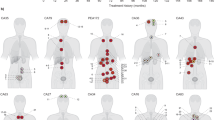
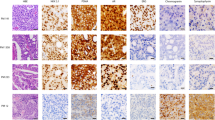
Abstract
Many studies have shown that primary prostate cancers are multifocal1,2,3 and are composed of multiple genetically distinct cancer cell clones4,5,6. Whether or not multiclonal primary prostate cancers typically give rise to multiclonal or monoclonal prostate cancer metastases is largely unknown, although studies at single chromosomal loci are consistent with the latter case. Here we show through a high-resolution genome-wide single nucleotide polymorphism and copy number survey that most, if not all, metastatic prostate cancers have monoclonal origins and maintain a unique signature copy number pattern of the parent cancer cell while also accumulating a variable number of separate subclonally sustained changes. We find no relationship between anatomic site of metastasis and genomic copy number change pattern. Taken together with past animal and cytogenetic studies of metastasis7 and recent single-locus genetic data in prostate and other metastatic cancers8,9,10, these data indicate that despite common genomic heterogeneity in primary cancers, most metastatic cancers arise from a single precursor cancer cell. This study establishes that genomic archeology of multiple anatomically separate metastatic cancers in individuals can be used to define the salient genomic features of a parent cancer clone of proven lethal metastatic phenotype.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Accession codes
Change history
07 July 2009
In the version of this article initially published, some of the sample identifiers were missing from Figure 1e. The error has been corrected in the HTML and PDF versions of the article.
References
Miller, G.J. & Cygan, J.M. Morphology of prostate cancer: the effects of multifocality on histological grade, tumor volume and capsule penetration. J. Urol. 152, 1709–1713 (1994).
Ruijter, E.T., van de Kaa, C.A., Schalken, J.A., Debruyne, F.M. & Ruiter, D.J. Histological grade heterogeneity in multifocal prostate cancer. Biological and clinical implications. J. Pathol. 180, 295–299 (1996).
Aihara, M., Wheeler, T.M., Ohori, M. & Scardino, P.T. Heterogeneity of prostate cancer in radical prostatectomy specimens. Urology 43, 60–66 (1994).
Cheng, L. et al. Evidence of independent origin of multiple tumors from patients with prostate cancer. J. Natl. Cancer Inst. 90, 233–237 (1998).
Macintosh, C.A., Stower, M., Reid, N. & Maitland, N.J. Precise microdissection of human prostate cancers reveals genotypic heterogeneity. Cancer Res. 58, 23–28 (1998).
Cheng, L. et al. Allelic imbalance in the clonal evolution of prostate carcinoma. Cancer 85, 2017–2022 (1999).
Fidler, I.J. & Talmadge, J.E. Evidence that intravenously derived murine pulmonary melanoma metastases can originate from the expansion of a single tumor cell. Cancer Res. 46, 5167–5171 (1986).
Kuukasjärvi, T. et al. Genetic heterogeneity and clonal evolution underlying development of asynchronous metastasis in human breast cancer. Cancer Res. 57, 1597–1604 (1997).
Sabatino, M. et al. Conservation of genetic alterations in recurrent melanoma supports the melanoma stem cell hypothesis. Cancer Res. 68, 122–131 (2008).
Mehra, R. et al. Characterization of TMPRSS2-ETS gene aberrations in androgen-independent metastatic prostate cancer. Cancer Res. 68, 3584–3590 (2008).
Tusher, V.G., Tibshirani, R. & Chu, G. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. USA 98, 5116–5121 (2001).
Zhu, Y. et al. A ground truth based comparative study on clustering of gene expression data. Front. Biosci. 13, 3839–3849 (2008).
Hastie, T.R., Tibshirani, T.R. & Friedman, J. The Elements of Statistical Learning: Data Mining, Inference, and Prediction, 415–431 (Springer, New York, 2001).
Wang, Z. et al. Optimized multilayer perceptrons for molecular classification and diagnosis using genomic data. Bioinformatics 22, 755–761 (2006).
Loog, M., Duin, R. & Haeb-Umbach, R. Multiclass linear dimension reduction by weighted pairwise fisher criteria. IEEE Trans. Pattern Anal. Mach. Intell. 23, 762–766 (2001).
Paget, S. The distribution of secondary growths in cancer of the breast. Lancet 133, 571–573 (1889).
Nowell, P.C. The clonal evolution of tumor cell populations. Science 194, 23–28 (1976).
Heim, S., Mandahl, N. & Mitelman, F. Genetic convergence and divergence in tumor progression. Cancer Res. 48, 5911–5916 (1988).
Eastham, J.A. et al. Association of p53 mutations with metastatic prostate cancer. Clin. Cancer Res. 1, 1111–1118 (1995).
Suzuki, H. et al. Interfocal heterogeneity of PTEN/MMAC1 gene alterations in multiple metastatic prostate cancer tissues. Cancer Res. 58, 204–209 (1998).
Bova, G.S., Chan-Tack, K. & LeCates, W.W. Lethal metastatic human prostate cancer: a review of the literature with emphasis on autopsy studies and characteristics of metastases in Prostate Cancer: Biology, Genetics, and New Therapeutics 39–60 (Humana Press, Totowa, New Jersey, 2000).
Powell, I.J. Epidemiology and pathophysiology of prostate cancer in African-American men. J. Urol. 177, 444–449 (2007).
Koivisto, P. et al. Androgen receptor gene amplification: a possible molecular mechanism for androgen deprivation therapy failure in prostate cancer. Cancer Res. 57, 314–319 (1997).
Miyoshi, Y. et al. Fluorescence in situ hybridization evaluation of c-myc and androgen receptor gene amplification and chromosomal anomalies in prostate cancer in Japanese patients. Prostate 43, 225–232 (2000).
Yegnasubramanian, S. et al. Hypermethylation of CpG islands in primary and metastatic human prostate cancer. Cancer Res. 64, 1975–1986 (2004).
Yegnasubramanian, S. et al. DNA hypomethylation arises later in prostate cancer progression than CpG island hypermethylation and contributes to metastatic tumor heterogeneity. Cancer Res. 68, 8954–8967 (2008).
Shah, R.B. et al. Androgen-independent prostate cancer is a heterogeneous group of diseases: lessons from a rapid autopsy program. Cancer Res. 64, 9209–9216 (2004).
Lengauer, C., Kinzler, K.W. & Vogelstein, B. Genetic instability in colorectal cancers. Nature 386, 623–627 (1997).
Vander Griend, D.J. et al. The role of CD133 in normal human prostate stem cells and malignant cancer-initiating cells. Cancer Res. 68, 9703–9711 (2008).
Ellis, W.J. et al. Detection and isolation of prostate cancer cells from peripheral blood and bone marrow. Urology 61, 277–281 (2003).
Vessella, R.L., Pantel, K. & Mohla, S. Tumor cell dormancy: an NCI workshop report. Cancer Biol. Ther. 6, 1496–1504 (2007).
Saramäki, O.R., Porkka, K.P., Vessella, R.L. & Visakorpi, T. Genetic aberrations in prostate cancer by microarray analysis. Int. J. Cancer 119, 1322–1329 (2006).
Kallioniemi, A. et al. Comparative genomic hybridization for molecular cytogenetic analysis of solid tumors. Science 258, 818–821 (1992).
Good, P.I. Permutation, Parametric and Bootstrap Tests of Hypotheses Ch. 3 (Springer, New York, 2005).
Kowalski, J., Pagano, M. & DeGruttola, V. A Nonparametric Test of Gene Region Heterogeneity Associated With Phenotype. J. Am. Stat. Assoc. 97, 398–408 (2002).
Acknowledgements
We thank the participating men and their families who suffered through metastatic prostate cancer and nonetheless gave the gift of participation so that others might benefit. We also thank V. Sinibaldi, T.B. Smyth and G.J. Mamo for oncologic and urologic clinical support, the Johns Hopkins Pathology Autopsy Service, including B. Crain and G. Hutchins, and A. Alkula, S. Kuivanen, T. Vilkkilä-Qwick, M. Vakkuri, D. Jay, X. Yi, S. H. Hahm, K. Jeffers Keiger, S. H. Chen, P. Powers, M. Taylor, C. Kang and Partek Customer Support for technical assistance. This work, commenced in 1994, was supported in part by Pirkanmaa Cancer Foundation, Maud Kuistila Foundation, Finnish Medical Foundation, the Medical Research Fund of Tampere University Hospital, Academy of Finland, Cancer Society of Finland, Reino Lahtikari Foundation, Sigrid Juselius Foundation, CaPCURE Foundation, John and Kathe Dyson, David Koch, the US National Institutes of Health National Cancer Institute (CA92234), the Prostate Cancer Research and Education Foundation, the US Department of Defense Congressionally Directed Prostate Cancer Research Program, the Grove Foundation and the American Cancer Society.
Author information
Authors and Affiliations
Contributions
W.L. performed and analyzed Affy6 assays and contributed to the manuscript and direction of study; S.L. performed and analyzed cCGH assays and contributed to the manuscript; S.K. performed statistical analysis of cCGH data and contributed to the manuscript; M.V. supervised statistical analysis of cCGH data and contributed to the manuscript; J.K. supervised and directed statistical analysis of cCGH and Affy6 data and contributed to the manuscript; G.Y. and L.C. performed statistical analysis of cCGH and Affy6 data and contributed to the manuscript; C.M.E. performed TMPRSS2-ERG transcript assays and contributed to the manuscript; M.A.E. and M.A.C. provided clinical support for the study; W.G.N. contributed to the manuscript and the analysis of chemotherapy data; S.Y. contributed to the manuscript and the direction of the study; J.L. contributed to the TMPRSS2-ERG transcript analysis and contributed to the manuscript; Y.W. supervised and directed cCGH and Affy6 data analysis and contributed to the manuscript; J.X. supervised Affy6 data collection and analysis and contributed to the direction of the study; W.B.I. supervised TMPRSS2-ERG transcript analysis and contributed to the manuscript and the direction of the study; T.V. supervised and interpreted cCGH data collection and contributed to the manuscript and the direction of the study; and G.S.B. founded and directed the autopsy project, directed the study, analyzed data and wrote the manuscript.
Corresponding author
Supplementary information
Supplementary Text and Figures
Supplementary Figs. 1–14, Supplementary Tables 1–11 and Supplementary Methods (PDF 902 kb)
Rights and permissions
About this article
Cite this article
Liu, W., Laitinen, S., Khan, S. et al. Copy number analysis indicates monoclonal origin of lethal metastatic prostate cancer. Nat Med 15, 559–565 (2009). https://doi.org/10.1038/nm.1944
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nm.1944
This article is cited by
-
Rapid assessment of 3-dimensional intra-tumor heterogeneity through cycling temperature capillary electrophoresis
BMC Research Notes (2023)
-
Focal prostate cancer therapy in the era of multiparametric MRI: a review of options and outcomes
Prostate Cancer and Prostatic Diseases (2023)
-
From molecular mechanisms of prostate cancer to translational applications: based on multi-omics fusion analysis and intelligent medicine
Health Information Science and Systems (2023)
-
Heterogeneity of the tumor immune microenvironment and clinical interventions
Frontiers of Medicine (2023)
-
Cancer origin tracing and timing in two high-risk prostate cancers using multisample whole genome analysis: prospects for personalized medicine
Genome Medicine (2023)