Abstract
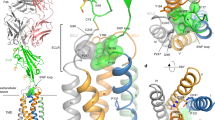
CD1 antigens bind a variety of self and foreign lipid and glycolipid antigens for presentation to CD1-restricted T cell receptors (TCRs). Here we report the crystal structure of human CD1a in complex with a sulfatide self antigen at a resolution of 2.15 Å. The lipid adopts an S-shaped conformation, with the sphingosine chain completely buried in the A′ pocket and the fatty acid chain emerging from the interface of the A′ pocket into the more exposed F′ pocket. The headgroup is anchored in the A′-F′ junction and protrudes into the F′ pocket for TCR recognition. Because the A′ pocket is narrow with a fixed terminus, it can act as a molecular 'ruler' to select alkyl chains of a particular length.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Accession codes
References
Calabi, F., Jarvis, J.M., Martin, L. & Milstein, C. Two classes of CD1 genes. Eur. J. Immunol. 19, 285–292 (1989).
Kawano, T. et al. CD1d-restricted and TCR-mediated activation of vα14 NKT cells by glycosylceramides. Science 278, 1626–1629 (1997).
Beckman, E.M. et al. CD1c restricts responses of mycobacteria-specific T cells. Evidence for antigen presentation by a second member of the human CD1 family. J. Immunol. 157, 2795–2803 (1996).
Porcelli, S., Morita, C.T. & Brenner, M.B. CD1b restricts the response of human CD4-8-T lymphocytes to a microbial antigen. Nature 360, 593–597 (1992).
Vincent, M.S. et al. CD1-dependent dendritic cell instruction. Nat. Immunol. 3, 1163–1168 (2002).
Rosat, J.P. et al. CD1-restricted microbial lipid antigen-specific recognition found in the CD8+ αβ T cell pool. J. Immunol. 162, 366–371 (1999).
Moody, D.B. et al. Structural requirements for glycolipid antigen recognition by CD1b-restricted T cells. Science 278, 283–286 (1997).
Shamshiev, A. et al. Self glycolipids as T-cell autoantigens. Eur. J. Immunol. 29, 1667–1675 (1999).
Shamshiev, A. et al. Presentation of the same glycolipid by different CD1 molecules. J. Exp. Med. 195, 1013–1021 (2002).
Moody, D.B. et al. CD1c-mediated T-cell recognition of isoprenoid glycolipids in Mycobacterium tuberculosis infection. Nature 404, 884–888 (2000).
Beckman, E.M. et al. Recognition of a lipid antigen by CD1-restricted αβ+ T cells. Nature 372, 691–694 (1994).
Moody, D.B. & Porcelli, S.A. Intracellular pathways of CD1 antigen presentation. Nat. Rev. Immunol. 3, 11–22 (2003).
Sugita, M. et al. Separate pathways for antigen presentation by CD1 molecules. Immunity 11, 743–752 (1999).
Moody, D.B. et al. Lipid length controls antigen entry into endosomal and nonendosomal pathways for CD1b presentation. Nat. Immunol. 3, 435–442 (2002).
Jackman, R.M. et al. The tyrosine-containing cytoplasmic tail of CD1b is essential for its efficient presentation of bacterial lipid antigens. Immunity 8, 341–351 (1998).
Chiu, Y.H. et al. Distinct subsets of CD1d-restricted T cells recognize self-antigens loaded in different cellular compartments. J. Exp. Med. 189, 103–110 (1999).
Sugita, M., van Der Wel, N., Rogers, R.A., Peters, P.J. & Brenner, M.B. CD1c molecules broadly survey the endocytic system. Proc. Natl. Acad. Sci. USA 97, 8445–8450 (2000).
Sugita, M. et al. Failure of trafficking and antigen presentation by CD1 in AP-3- deficient cells. Immunity 16, 697–706 (2002).
Porcelli, S. et al. Recognition of cluster of differentiation 1 antigens by human CD4-CD8-cytolytic T lymphocytes. Nature 341, 447–450 (1989).
Zeng, Z. et al. Crystal structure of mouse CD1: an MHC-like fold with a large hydrophobic binding groove. Science 277, 339–345 (1997).
Gadola, S.D. et al. Structure of human CD1b with bound ligands at 2.3 Å, a maze for alkyl chains. Nat. Immunol. 3, 721–726 (2002).
Jackson, M.R., Song, E.S., Yang, Y. & Peterson, P.A. Empty and peptide-containing conformers of class I major histocompatibility complex molecules expressed in Drosophila melanogaster cells. Proc. Natl. Acad. Sci. USA 89, 12117–12121 (1992).
Matsumura, M., Saito, Y., Jackson, M.R., Song, E.S. & Peterson, P.A. In vitro peptide binding to soluble empty class I major histocompatibility complex molecules isolated from transfected Drosophila melanogaster cells. J. Biol. Chem. 267, 23589–23595 (1992).
Matsumura, M., Fremont, D.H., Peterson, P.A. & Wilson, I.A. Emerging principles for the recognition of peptide antigens by MHC class I molecules. Science 257, 927–934 (1992).
Madden, D.R. The three-dimensional structure of peptide-MHC complexes. Annu. Rev. Immunol. 13, 587–622.(1995).
O×Brien, J.S., Fillerup, D.L. & Mead, J.F. Quantification and fatty acid and fatty aldehyde composition of ethanolamine, choline, and serine glycerophosphatides in human cerebral grey and white matter. J. Lipid. Res. 5, 329–338 (1964).
Nicholls, A., Sharp, K.A. & Honig, B. Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins Struct. Funct. Genet. 11, 281–296 (1991).
O'Callaghan, C.A. et al. Structural features impose tight peptide binding specificity in the nonclassical MHC molecule HLA-E. Mol. Cell. 1, 531–541 (1998).
Khan, A.R., Baker, B.M., Ghosh, P., Biddison, W.E. & Wiley, D.C. The structure and stability of an HLA-A*0201/octameric tax peptide complex with an empty conserved peptide-N-terminal binding site. J. Immunol. 164, 6398–6405 (2000).
Speir, J.A., Abdel-Motal, U.M., Jondal, M. & Wilson, I.A. Crystal structure of an MHC class I presented glycopeptide that generates carbohydrate-specific CTL. Immunity 10, 51–61 (1999).
Lerche, M.H., Kragelund, B.B., Bech, L.M. & Poulsen, F.M. Barley lipid-transfer protein complexed with palmitoyl CoA: the structure reveals a hydrophobic binding site that can expand to fit both large and small lipid-like ligands. Structure 5, 291–306 (1997).
Young, A.C. et al. Structural studies on human muscle fatty acid binding protein at 1.4 Å resolution: binding interactions with three C18 fatty acids. Structure 2, 523–534 (1994).
Manolova, V., Hirabayashi, Y., Mori, L. & Libero, G.D. CD1a and CD1b surface expression is independent from de novo synthesized glycosphingolipids. Eur. J. Immunol. 33, 29–37 (2003).
Sidobre, S. & Kronenberg, M. CD1 tetramers: a powerful tool for the analysis of glycolipid-reactive T cells. J. Immunol. Methods 268, 107–121 (2002).
Otwinowski, Z. & Minor, W. HKL: Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997).
Vagin, A.A. & Teplyakov, A. MOLREP:an automated program for molecular replacement. J. Appl. Crystallogr. 30, 1022–1025 (1997).
Brünger, A.T. et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D54, 905–921 (1998).
Pannu, N.S. & Read, R.J. Improved structure refinement through maximum likelyhood. Acta Crystallogr. A52, 659–668 (1996).
Brünger, A.T. Free R value: a novel statistical quantity for assessing the accuracy of crystal structures. Nature 355, 472–475 (1992).
Jones, T.A., Cowan, S., Zou, J.Y. & Kjeldgaard, M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A47, 110–119 (1991).
Kleywegt, G.J. & Jones, T.A. Databases in protein crystallography. Acta Crystallogr. D54, 1119–1131 (1998).
Murshudov, G.N., Vagin, A.A. & Dodson, E.J. Refinement of macromolecular structures by the maximum likelihood method. Acta Crystallogr. D53, 240–255 (1997).
CCP4. Collaborative Computational Project, Number 4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D50, 760–763 (1994).
Winn, M.D., Isupov, M.N. & Murshudov, G.N. Use of TLS parameters to model anisotropic displacements in macromolecular refinement. Acta Crystallogr. D57, 122–133 (2001).
Laskowski, R.A., MacArthur, M.W., Moss, D.S. & Thornton, J.M. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26, 283–291 (1993).
Connolly, M.L. The molecular surface package. J. Mol. Graph. 11, 139–141 (1993).
Gelin, B.R. & Karplus, M. Side-chain torsional potentials: effect of dipeptide, protein, and solvent environment. Biochemistry 18, 1256–1268 (1979).
Sheriff, S., Hendrickson, W.A. & Smith, J.L. Structure of myohemerythrin in the azidomet state at 1.7/1.3 Å resolution. J. Mol. Biol. 197, 273–296 (1987).
Kraulis, P.J. MOLSCRIPT: a program to produce both detailed and schematic plots of proteins. J. Appl. Crystallogr. 24, 946–950 (1991).
Merritt, E.A. & Bacon, D.J. Raster3D: Photorealistic molecular graphics. Methods Enzymol. 277, 505–524 (1997).
Esnouf, R.M. An extensively modified version of Molscript that includes greatly enhanced coloring capabilities. J. Mol. Graph. Model. 15, 132–134 (1997).
Howlin, B., Butler, D.S., Moss, D.S., Harris, G.W. & Driessen, H.P.C. TLSANL: TLS parameter analysis program for segmented anisotropic refinement of macromolecular structures. J. Appl. Crystallogr. 26, 622–624 (1993).
Acknowledgements
We thank the staff of the Advanced Light Source BL 5.0.1 for support with data collection; M. Crispin, R. Stanfield, A. Heine, J. Stevens, J. Luz, N. Larsen, J. Kelly and B. Moody for discussions; X. Dai for assistance on synchrotron trips; S. Ferguson for technical assistance; R. Stefanko and M. Wallace for generating the CD1a construct; and E. Grant and M. Brenner for the CD1a cDNA. We acknowledge Syrrx for the use of their crystallization robot (initial CD1a crystallization trials). This study was supported by National Institutes of Health grants GM62116 (I.A.W.), CA58896 (I.A.W.) and AI53725 (L.T.), postdoctoral fellowships from the Deutsche Forschungsgemeinschaft and the Skaggs Institute for Chemical Biology (D.M.Z.).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Zajonc, D., Elsliger, M., Teyton, L. et al. Crystal structure of CD1a in complex with a sulfatide self antigen at a resolution of 2.15 Å. Nat Immunol 4, 808–815 (2003). https://doi.org/10.1038/ni948
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ni948
This article is cited by
-
Structural, in silico, and functional analysis of a Disabled-2-derived peptide for recognition of sulfatides
Scientific Reports (2020)
-
CD1c caves in on lipids
Nature Immunology (2018)
-
Conservation of sequence motifs suggests that the nonclassical MHC class I lineages CD1/PROCR and UT were established before the emergence of tetrapod species
Immunogenetics (2018)
-
T cell autoreactivity directed toward CD1c itself rather than toward carried self lipids
Nature Immunology (2018)
-
CD1: A Singed Cat of the Three Antigen Presentation Systems
Archivum Immunologiae et Therapiae Experimentalis (2017)