Abstract
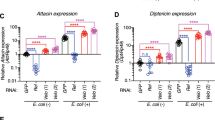
The Drosophila immune system discriminates between different classes of infectious microbes and responds with pathogen-specific defense reactions through selective activation of the Toll and the immune deficiency (Imd) signaling pathways. The Toll pathway mediates most defenses against Gram-positive bacteria and fungi, whereas the Imd pathway is required to resist infection by Gram-negative bacteria. The bacterial components recognized by these pathways remain to be defined. Here we report that Gram-negative diaminopimelic acid–type peptidoglycan is the most potent inducer of the Imd pathway and that the Toll pathway is predominantly activated by Gram-positive lysine-type peptidoglycan. Thus, the ability of Drosophila to discriminate between Gram-positive and Gram-negative bacteria relies on the recognition of specific forms of peptidoglycan.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Medzhitov, R. & Janeway, C.A. Jr. Innate immunity: the virtues of a nonclonal system of recognition. Cell 91, 295–298 (1997).
Tzou, P., De Gregorio, E. & Lemaitre, B. How Drosophila combats microbial infection: a model to study innate immunity and host-pathogen interactions. Curr. Opin. Microbiol. 5, 102–110 (2002).
Hultmark, D. Drosophila immunity: paths and patterns. Curr. Opin. Immunol. 15, 12–19 (2003).
Khush, R.S., Leulier, F. & Lemaitre, B. Drosophila immunity: two paths to NF-κB. Trends Immunol. 22, 260–264 (2001).
De Gregorio, E., Spellman, P.T., Tzou, P., Rubin, G.M. & Lemaitre, B. The Toll and Imd pathways are the major regulators of the immune response in Drosophila. EMBO J. 21, 2568–2579 (2002).
Boutros, M., Agaisse, H. & Perrimon, N. Sequential activation of signaling pathways during innate immune response in Drosophila. Dev. Cell 3, 711–722 (2002).
Lemaitre, B., Reichhart, J. & Hoffmann, J. Drosophila host defense: differential induction of antimicrobial peptide genes after infection by various classes of microorganisms. Proc. Natl. Acad. Sci. USA 94, 14614–14619 (1997).
Leulier, F., Rodriguez, A., Khush, R.S., Abrams, J.M. & Lemaitre, B. The Drosophila caspase Dredd is required to resist Gram-negative bacterial infection. EMBO Rep. 1, 353–358 (2000).
Rutschmann, S. et al. The Rel protein DIF mediates the antifungal but not the antibacterial host defense in Drosophila. Immunity 12, 569–580 (2000).
Rutschmann, S. et al. Role of Drosophila IKKγ in a Toll-independent antibacterial immune response. Nat. Immunol. 1, 342–347 (2000).
Vidal, S. et al. Mutations in the Drosophila dTAK1 gene reveal a conserved function for MAPKKKs in the control of rel/NF-κB–dependent innate immune responses. Genes Dev. 15, 1900–1912 (2001).
Hedengren, M. et al. Relish, a central factor in the control of humoral but not cellular immunity in Drosophila. Mol. Cell 4, 827–837 (1999).
Yoshida, H., Kinoshita, K. & Ashida, M. Purification of a peptidoglycan recognition protein from hemolymph of the silkworm, Bombyx mori. J. Biol. Chem. 271, 13854–13860. (1996).
Kang, D., Liu, G., Lundstrom, A., Gelius, E. & Steiner, H. A peptidoglycan recognition protein in innate immunity conserved from insects to humans. Proc. Natl. Acad. Sci. USA 95, 10078–10082 (1998).
Werner, T. et al. A family of peptidoglycan recognition proteins in the fruit fly Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 97, 13772–13777 (2000).
Michel, T., Reichhart, J.M., Hoffmann, J.A. & Royet, J. Drosophila Toll is activated by Gram-positive bacteria through a circulating peptidoglycan recognition protein. Nature 414, 756–759 (2001).
Choe, K.M., Werner, T., Stoven, S., Hultmark, D. & Anderson, K.V. Requirement for a peptidoglycan recognition protein (PGRP) in Relish activation and antibacterial immune responses in Drosophila. Science 296, 359–362 (2002).
Gottar, M. et al. The Drosophila immune response against Gram-negative bacteria is mediated by a peptidoglycan recognition protein. Nature 416, 640–644 (2002).
Ramet, M., Manfruelli, P., Pearson, A., Mathey-Prevot, B. & Ezekowitz, R.A. Functional genomic analysis of phagocytosis and identification of a Drosophila receptor for E. coli. Nature 416, 644–648 (2002).
Takehana, A. et al. Overexpression of a pattern-recognition receptor, peptidoglycan- recognition protein-LE, activates imd/relish-mediated antibacterial defense and the prophenoloxidase cascade in Drosophila larvae. Proc. Natl. Acad. Sci. USA 99, 13705–13710 (2002).
Akira, S., Takeda, K. & Kaisho, T. Toll-like receptors: critical proteins linking innate and acquired immunity. Nat. Immunol. 2, 675–680 (2001).
Janeway, C.A. Jr. & Medzhitov, R. Innate immune recognition. Annu. Rev. Immunol. 20, 197–216 (2002).
Samakovlis, C., Asling, B., Boman, H., Gateff, E. & Hultmark, D. In vitro induction of cecropin genes: an immune response in a Drosophila blood cell line. Biochem. Biophys. Res. Commun. 188, 1169–1175 (1992).
Imler, J.L., Tauszig, S., Jouanguy, E., Forestier, C. & Hoffmann, J.A. LPS-induced immune response in Drosophila. J. Endotoxin Res. 6, 459–462 (2000).
Hirschfeld, M., Ma, Y., Weis, J.H., Vogel, S.N. & Weis, J.J. Cutting edge: repurification of lipopolysaccharide eliminates signaling through both human and murine toll-like receptor 2. J. Immunol. 165, 618–622 (2000).
Glauner, B., Holtje, J.V. & Schwarz, U. The composition of the murein of Escherichia coli. J. Biol. Chem. 263, 10088–10095 (1988).
Mengin-Lecreulx, D. & van Heijenoort, J. Effect of growth conditions on peptidoglycan content and cytoplasmic steps of its biosynthesis in Escherichia. J. Bacteriol. 163, 208–212 (1985).
Lemaitre, B., Nicolas, E., Michaut, L., Reichhart, J. & Hoffmann, J. The dorsoventral regulatory gene cassette spätzle/Toll/cactus controls the potent antifungal response in Drosophila adults. Cell 86, 973–983 (1996).
Girardin, S.E. et al. Nod2 is a general sensor of peptidoglycan through muramyl dipeptide (MDP) detection. J. Biol. Chem. 278, 8869–8872 (2003).
Inohara, N. et al. Host recognition of bacterial muramyl dipeptide mediated through NOD2: implications for Crohn's disease. J. Biol. Chem. 278, 5509–5512 (2003).
Ashida, M., Ishizaki, Y. & Iwahana, H. Activation of pro-phenoloxidase by bacterial cell walls or β-1,3-glucans in plasma of the silkworm, Bombyx mori. Biochem. Biophys. Res. Commun 113, 562–568 (1983).
Yoshida, H. & Ashida, M. Microbial activation of two serine enzymes and prophenoloxidase in the plasma fraction of hemolymph of the silkworm, Bombyx mori. Insect Biochem. 16, 539–545 (1986).
Schleifer, K.H. & Kandler, O. Peptidoglycan types of bacterial cell walls and their taxonomic implications. Bacteriol. Rev. 36, 407–477 (1972).
Park, J.T. Why does Escherichia coli recycle its cell wall peptides? Mol. Microbiol. 17, 421–426 (1995).
Pye, A. Microbial activation of prophenoloxidase from immune insect larvae. Nature 251, 610–613 (1974).
Iketani, M., Nishimura, H., Akayama, K., Yamano, Y. & Morishima, I. Minimum structure of peptidoglycan required for induction of antibacterial protein synthesis in the silkworm, Bombyx mori. Insect Biochem. Mol. Biol. 29, 19–24 (1999).
Takeuchi, O. et al. Differential roles of TLR2 and TLR4 in recognition of Gram-negative and Gram-positive bacterial cell wall components. Immunity 11, 443–451 (1999).
Tzou, P., Meister, M. & Lemaitre, B. Methods for studying infection and immunity in Drosophila. Methods Microbiol. 31, 507–529 (2002).
Aussel, L., Brisson, J.R., Perry, M.B. & Caroff, M. Structure of the lipid A of Bordetella hinzii ATCC 51730. Rapid Commun. Mass Spectrom. 14, 595–599 (2000).
Karibian, D., Deprun, C. & Caroff, M. A comparison of the Lipid A of several Salmonella and Escherichia strains by 252Cf plasma desorption mass spectrometry. J. Bacteriol. 2988–2993 (1993).
Shands, J.W. Jr. & Chun, P.W. The dispersion of Gram-negative lipopolysaccharide by deoxycholate. Subunit molecular weight. J. Biol. Chem. 255, 1221–1226 (1980).
Datsenko, K.A. & Wanner, B.L. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97, 6640–6645 (2000).
Acknowledgements
We thank E. De Gregorio and F. Boccard for reading the manuscript; N. Vodovar for technical help; R. Chaby, D. Philpott and S. Girardin for discussions, and J. Royet, D. Ferrandon and K.V. Anderson for fly stocks. This work was supported by the Centre National de la Recherche Scientifique and grants from Association pour la Recherche sur la Cancer, Programme de Recherche Fondamentale en Microbiologie et Maladies Infectieuses et Parasitaires and the Agence Nationale de Recherche sur le Sida.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Leulier, F., Parquet, C., Pili-Floury, S. et al. The Drosophila immune system detects bacteria through specific peptidoglycan recognition. Nat Immunol 4, 478–484 (2003). https://doi.org/10.1038/ni922
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ni922
This article is cited by
-
Prenatal Programming of Monocyte Chemotactic Protein-1 Signaling in Autism Susceptibility
Molecular Neurobiology (2024)
-
Transcriptome analysis of the endangered dung beetle Copris tripartitus (Coleoptera: Scarabaeidae) and characterization of genes associated to immunity, growth, and reproduction
BMC Genomics (2023)
-
Drice restrains Diap2-mediated inflammatory signalling and intestinal inflammation
Cell Death & Differentiation (2022)
-
Characterization of PGRP-LB and immune deficiency in the white-backed planthopper Sogatella furcifera (Hemiptera: Delphacidae)
Applied Entomology and Zoology (2022)
-
Sensing microbial infections in the Drosophila melanogaster genetic model organism
Immunogenetics (2022)