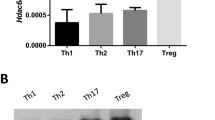
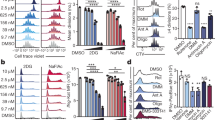
Abstract
Naïve T cells differentiate into effector cells upon stimulation with antigen, a process that is accompanied by changes in the chromatin structure of effector cytokine genes. Using histone acetylation to evaluate these changes, we showed that T cell receptor (TCR) stimulation results in early activation of the genes encoding both interleukin 4 and interferon-γ. We found that continued culture in the presence of polarizing cytokines established a selective pattern of histone acetylation on both cytokine genes; this correlated with restricted access of the transcription factor NFAT1 to these gene regulatory regions as well as mutually exclusive gene expression by the differentiated T cells. Our data point to a biphasic process in which cytokine-driven signaling pathways maintain and reinforce chromatin structural changes initiated by the TCR. This process ensures that cytokine genes remain accessible to the relevant transcription factors and promotes functional cooperation of the inducible transcription factor NFAT with lineage-specific transcription factors such as GATA-3 and T-bet.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Abbas, A.K., Murphy, K.M. & Sher, A. Functional diversity of helper T lymphocytes. Nature 383, 787–793 (1996).
Wurster, A.L., Tanaka, T. & Grusby, M.J. The biology of Stat4 and Stat6. Oncogene 19, 2577–2584 (2000).
Murphy, K.M. et al. Signaling and transcription in T helper development. Annu. Rev. Immunol. 18, 451–494 (2000).
Glimcher, L.H. & Murphy, K.M. Lineage commitment in the immune system: the T helper lymphocyte grows up. Genes Dev. 14, 1693–1711 (2000).
Reiner, S.L. Helper T cell differentiation, inside and out. Curr. Opin. Immunol. 13, 351–355 (2001).
Avni, O. & Rao, A. T cell differentiation: a mechanistic view. Curr. Opin. Immunol. 12, 654–659 (2000).
O'Garra, A. & Arai, N. The molecular basis of T helper 1 and T helper 2 cell differentiation. Trends Cell Biol. 10, 542–550 (2000).
Lee, D., Agarwal, S. & Rao, A. Th2 lineage commitment and efficient IL-4 production involves extended demethylation of the IL-4 gene. Immunity 16, 649–660 (2002).
Agarwal, S. & Rao, A. Modulation of chromatin structure regulates cytokine gene expression during T cell differentiation. Immunity 9, 765–775 (1998).
Takemoto, N. et al. Th2-specific DNAse I-hypersensitive sites in the murine IL-13 and IL-4 intergenic region. Int. Immunol. 10, 1981–1985 (1998).
Agarwal, S., Avni, O. & Rao, A. Cell-type-restricted binding of the transcription factor NFAT to a distal IL-4 enhancer in vivo. Immunity 12, 643–652 (2000).
Mohrs, M. et al. Deletion of a coordinate regulator of type 2 cytokine expression in mice. Nature Immunol. 2, 842–847 (2001).
Loots, G.G. et al. Identification of a coordinate regulator of interleukins 4, 13, and 5 by cross-species sequence comparisons. Science 288, 136–140 (2000).
Solymar, D.C., Agarwal, S., Bassing, C.H., Alt, F.W. & Rao, A. A 3' enhancer in the IL-4 gene regulates cytokine production by Th2 cells and mast cells. Immunity, (in the press, 2002).
Jenuwein, T. & Allis, C.D. Translating the histone code. Science 293, 1074–1080 (2001).
Rice, J.C. & Allis, C.D. Histone methylation versus histone acetylation: new insights into epigenetic regulation. Curr. Opin. Cell Biol. 13, 263–273 (2001).
Strahl, B.D. & Allis, C.D. The language of covalent histone modifications. Nature 403, 41–45 (2000).
Cheung, P., Allis, C.D. & Sassone-Corsi, P. Signaling to chromatin through histone modifications. Cell 103, 263–271 (2000).
Roth, S.Y., Denu, J.M. & Allis, C.D. Histone Acetyltransferases. Annu. Rev. Biochem. 70, 81–120 (2001).
Lederer, J.A. et al. Cytokine transcriptional events during helper T cell subset differentiation. J. Exp. Med. 184, 397–406 (1996).
Kurata, H., Lee, H., O'Garra, A. & Arai, N. Ectopic expression of activated Stat6 induces the expression of Th2-specific cytokines and transcription factors in developing Th1 cells. Immunity 11, 677–688 (1999).
Lee, H.J. et al. GATA-3 induces T helper cell type 2 (Th2) cytokine expression and chromatin remodeling in committed Th1 cells. J. Exp. Med. 192, 105–116 (2000).
Ouyang, W. et al. Stat6-independent GATA-3 autoactivation directs IL-4-independent Th2 development and commitment. Immunity 12, 27–37 (2000).
Ho, I.-C., Hodge, M.R., Rooney, J.W. & Glimcher, L.H. The proto-oncogene c-maf is responsible for tissue-specific expression of interleukin-4. Cell 85, 973–983 (1996).
Macian, F., Garcia-Rodriguez, C. & Rao, A. Gene expression elicited by NFAT in the presence or absence of cooperative recruitment of Fos and Jun. EMBO J. 19, 4783–4795 (2000).
Szabo, S.J. et al. A novel transcription factor, T-bet, directs Th1 lineage commitment. Cell 100, 655–669 (2000).
Szabo, S.J. et al. Distinct effects of T-bet in TH1 lineage commitment and IFN-γ production in CD4 and CD8 T cells. Science 295, 338–342 (2002).
Kamogawa, Y., Minasi, L.-A., Carding, S., Bottomly, K. & Flavell, R. The relationship of IL4 and IFNγ-producing T cells studied by lineage ablation of IL4-producing cells. Cell 75, 985–995 (1993).
Croft, M. & Swain, S.L. Recently activated naïve CD4 T cells can help resting B cells, and can produce sufficient autocrine IL-4 to drive differentiation to secretion of T helper 2-type cytokines. J. Immunol. 154, 4269–4282 (1995).
Noben-Trauth, N., Hu-Li, J. & Paul, W.E. Conventional, naïve CD4+ T cells provide an initial source of IL-4 during Th2 differentiation. J. Immunol. 165, 3620–3625 (2000).
Finkelman, F.D. et al. Stat6 regulation of in vivo IL-4 responses. J. Immunol. 164, 2303–2310 (2000).
Grogan, J.L. et al. Early transcription and silencing of cytokine genes underlie polarization of T helper cell subsets. Immunity 14, 205–215 (2001).
Zhao, K. et al. Rapid and phosphoinositol-dependent binding of the SWI/SNF-like BAF complex to chromatin after T lymphocyte receptor signaling. Cell 95, 625–636 (1999).
Garcia-Rodriguez, C. & Rao, A. Nuclear factor of activated T cells (NFAT)-dependent transactivation regulated by the coactivators p300/CREB-binding protein (CBP). J. Exp. Med. 187, 2031–2036 (1998).
Avots, A. et al. CBP/p300 integrates Raf/Rac-signaling pathways in the transcriptional induction of NF-ATc during T cell activation. Immunity 10, 515–524 (1999).
Lachner, M., O'Carroll, D., Rea, S., Mechtler, K. & Jenuwein, T. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature 410, 116–120 (2001).
Brown, K.E. et al. Association of transcriptionally silent genes with Ikaros complexes at centromeric heterochromatin. Cell 91, 845–854 (1997).
Kaplan, M.H., Schindler, U., Smiley, S. & Grusby, M. Stat6 is required for mediating responses to IL-4 and for development of TH2 cells. Immunity 4, 313–319 (1996).
Shimoda, K. et al. Lack of IL-4-induced Th2 response and IgE class switching in mice with disrupted Stat6 gene. Nature 380, 630–633 (1996).
Takeda, K. et al. Essential role of Stat6 in IL-4 signalling. Nature 380, 627–630 (1996).
Kopf, M. et al. Disruption of the IL-4 gene blocks Th2 cytokine responses. Nature 362, 245–248 (1993).
Jankovic, D. et al. Single cell analysis reveals that IL-4 receptor/Stat6 signaling is not required for the in vivo or in vitro development of CD4+ lymphocytes with a Th2 cytokine profile. J. Immunol. 164, 3047–3055 (2000).
Gingras, S., Simard, J., Groner, B. & Pfitzner, E. p300/CBP is required for transcriptional induction by interleukin-4 and interacts with Stat6. Nucleic Acids Res. 27, 2722–2729 (1999).
Zhang, D. et al. Inhibition of allergic inflammation in a murine model of asthma by expression of a dominant-negative mutant of GATA-3. Immunity 11, 473–482 (1999).
Zheng, W. & Flavell, R.A. The transcription factor GATA-3 is necessary and sufficient for Th2 cytokine gene expression in CD4 T cells. Cell 89, 587–596 (1997).
Ouyang, W. et al. Inhibition of Th1 development mediated by GATA-3 through an IL-4-independent mechanism. Immunity 9, 745–755 (1998).
Takemoto, N. et al. Cutting edge: chromatin remodeling at the IL-4/IL-13 intergenic regulatory region for Th2-specific cytokine gene cluster. J. Immunol. 165, 6687–6691 (2000).
Henkel, G. & Brown, M.A. PU.1 and GATA: components of a mast cell-specific interleukin 4 intronic enhancer. Proc. Natl. Acad. Sci. USA 91, 7737–7741 (1994).
Ranganath, S. et al. GATA-3-dependent enhancer activity in IL-4 gene regulation. J. Immunol. 161, 3822–3826 (1998).
Lee, G.R., Fields, P.E. & Flavell, R.A. Regulation of IL-4 gene expression by distal regulatory elements and GATA-3 at the chromatin level. Immunity 14, 447–459 (2001).
Murphy, K.M., Heimberger, A.B. & Loh, D.Y. Induction by antigen of intrathymic apoptosis of CD4+CD8+TCRlo thymocytes in vivo. Science 250, 1720–1723 (1990).
Tepper, R.I., Pattengale, P.K. & Leder, P. Murine interleukin-4 displays potent anti-tumor activity in vivo. Cell 57, 503–512 (1989).
Parekh, B. & Maniatis, T. Virus infection leads to localized hyperacetylation of histones H3 and H4 at the IFN-β promoter. Mol. Cell 3, 125–129 (1999).
Wang, D., McCaffrey, P. & Rao, A. The cyclosporin-sensitive transcription factor NFATp is expressed in several classes of cells in the immune system. Ann. NY Acad. Sci. 766, 182–194 (1995).
Luo, C. et al. Recombinant NFAT1 (NFATp) is regulated by calcineurin in T cells and mediates transcription of several cytokine genes. Mol. Cell Biol. 16, 3955–3966 (1996).
Acknowledgements
We thank members of the laboratory for critical reading of the manuscript and valuable discussions. Supported by grants from the National Institutes of Health (to A. R. and L. H. G.), a gift from the G. Harold and Leila Y. Mathers Charitable Foundation (to L. H. G.), the Cancer Research Institute (to O. A.), the National Institutes of Health (to D. L.) and the Leukemia Society and a grant from the Burroughs Wellcome Fund (to S. J. S.).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Avni, O., Lee, D., Macian, F. et al. TH cell differentiation is accompanied by dynamic changes in histone acetylation of cytokine genes. Nat Immunol 3, 643–651 (2002). https://doi.org/10.1038/ni808
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ni808
This article is cited by
-
Per-cell histone acetylation is associated with terminal differentiation in human T cells
Clinical Epigenetics (2024)
-
Epigenetic reprogramming of T cells: unlocking new avenues for cancer immunotherapy
Cancer and Metastasis Reviews (2024)
-
Protean role of epigenetic mechanisms and their impact in regulating the Tregs in TME
Cancer Gene Therapy (2022)
-
Repositioning TH cell polarization from single cytokines to complex help
Nature Immunology (2021)
-
Scientific divagations: from signaling and transcription to chromatin changes in T cells
Nature Immunology (2020)