Abstract
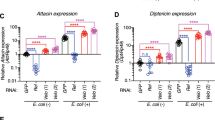
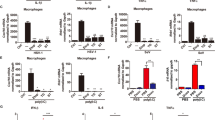
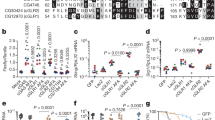
During a genome-wide screen with RNA-mediated interference, we isolated CG8580 as a gene involved in the innate immune response of Drosophila melanogaster. CG8580, which we called Akirin, encoded a protein that acted in parallel with the NF-κB transcription factor downstream of the Imd pathway and was required for defense against Gram-negative bacteria. Akirin is highly conserved, and the human genome contains two homologs, one of which was able to rescue the loss-of-function phenotype in drosophila cells. Akirins were strictly localized to the nucleus. Knockout of both Akirin homologs in mice showed that one had an essential function downstream of the Toll-like receptor, tumor necrosis factor and interleukin (IL)-1β signaling pathways leading to the production of IL-6. Thus, Akirin is a conserved nuclear factor required for innate immune responses.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Change history
11 January 2008
In the version of this article initially published, the bars for the LPS samples in Figure 6b are incorrect. The correct data are presented here.
11 December 2009
In addition, an incorrect accession code was given for Xenopus laevis Akirin1; the correct code is NP_001089245. These errors have been corrected in the HTML and PDF versions of the article.
References
Akira, S., Uematsu, S. & Takeuchi, O. Pathogen recognition and innate immunity. Cell 124, 783–801 (2006).
Brennan, C.A. & Anderson, K.V. Drosophila: the genetics of innate immune recognition and response. Annu. Rev. Immunol. 22, 457–483 (2004).
Hoffmann, J.A. The immune response of Drosophila. Nature 426, 33–38 (2003).
Lemaitre, B. & Hoffmann, J. The host defense of Drosophila melanogaster. Annu. Rev. Immunol. 25, 697–743 (2007).
Hoffmann, J.A. & Reichhart, J.M. Drosophila innate immunity: an evolutionary perspective. Nat. Immunol. 3, 121–126 (2002).
Lemaitre, B., Nicolas, E., Michaut, L., Reichhart, J.M. & Hoffmann, J.A. The dorsoventral regulatory gene cassette spatzle/Toll/cactus controls the potent antifungal response in Drosophila adults. Cell 86, 973–983 (1996).
Weber, A.N. et al. Binding of the Drosophila cytokine Spatzle to Toll is direct and establishes signaling. Nat. Immunol. 4, 794–800 (2003).
Ip, Y. et al. Dif, a dorsal-related gene that mediates an immune response in Drosophila. Cell 75, 753–763 (1993).
Rutschmann, S. et al. The Rel protein DIF mediates the antifungal but not the antibacterial host defense in Drosophila. Immunity 12, 569–580 (2000).
Tauszig-Delamasure, S., Bilak, H., Capovilla, M., Hoffmann, J.A. & Imler, J.L. Drosophila MyD88 is required for the response to fungal and Gram-positive bacterial infections. Nat. Immunol. 3, 91–97 (2002).
Stoven, S. et al. Caspase-mediated processing of the Drosophila NF-κB factor Relish. Proc. Natl. Acad. Sci. USA 100, 5991–5996 (2003).
Silverman, N. et al. A Drosophila IκB kinase complex required for Relish cleavage and antibacterial immunity. Genes Dev. 14, 2461–2471 (2000).
Dushay, M.S., Asling, B. & Hultmark, D. Origins of immunity: Relish, a compound Rel-like gene in the antibacterial defense of Drosophila. Proc. Natl. Acad. Sci. USA 93, 10343–10347 (1996).
Mak, T.W. & Yeh, W.C. Signaling for survival and apoptosis in the immune system. Arthritis Res. 4, S243–S252 (2002).
Choe, K.M., Werner, T., Stoven, S., Hultmark, D. & Anderson, K.V. Requirement for a peptidoglycan recognition protein (PGRP) in Relish activation and antibacterial immune responses in Drosophila. Science 296, 359–362 (2002).
Gottar, M. et al. The Drosophila immune response against Gram-negative bacteria is mediated by a peptidoglycan recognition protein. Nature 416, 640–644 (2002).
Leulier, F. et al. The Drosophila immune system detects bacteria through specific peptidoglycan recognition. Nat. Immunol. 4, 478–484 (2003).
Kaneko, T. et al. Monomeric and polymeric gram-negative peptidoglycan but not purified LPS stimulate the Drosophila IMD pathway. Immunity 20, 637–649 (2004).
Mellroth, P. et al. Ligand-induced dimerization of Drosophila peptidoglycan recognition proteins in vitro. Proc. Natl. Acad. Sci. USA 102, 6455–6460 (2005).
Takehana, A. et al. Overexpression of a pattern-recognition receptor, peptidoglycan-recognition protein-LE, activates imd/relish-mediated antibacterial defense and the prophenoloxidase cascade in Drosophila larvae. Proc. Natl. Acad. Sci. USA 99, 13705–13710 (2002).
Choe, K.M., Lee, H. & Anderson, K.V. Drosophila peptidoglycan recognition protein LC (PGRP-LC) acts as a signal-transducing innate immune receptor. Proc. Natl. Acad. Sci. USA 102, 1122–1126 (2005).
Georgel, P. et al. Drosophila immune deficiency (IMD) is a death domain protein that activates antibacterial defense and can promote apoptosis. Dev. Cell 1, 503–514 (2001).
Kelliher, M.A. et al. The death domain kinase RIP mediates the TNF-induced NF-κB signal. Immunity 8, 297–303 (1998).
Leulier, F., Rodriguez, A., Khush, R.S., Abrams, J.M. & Lemaitre, B. The Drosophila caspase Dredd is required to resist gram-negative bacterial infection. EMBO Rep. 1, 353–358 (2000).
Naitza, S. et al. The Drosophila immune defense against gram-negative infection requires the death protein dFADD. Immunity 17, 575–581 (2002).
Zhuang, Z.H. et al. Drosophila TAB2 is required for the immune activation of JNK and NF-κB. Cell. Signal. 18, 964–970 (2006).
Gesellchen, V., Kuttenkeuler, D., Steckel, M., Pelte, N. & Boutros, M. An RNA interference screen identifies Inhibitor of Apoptosis Protein 2 as a regulator of innate immune signalling in Drosophila. EMBO Rep. 6, 979–984 (2005).
Vidal, S. et al. Mutations in the Drosophila dTAK1 gene reveal a conserved function for MAPKKKs in the control of rel/NF-κB-dependent innate immune responses. Genes Dev. 15, 1900–1912 (2001).
Rutschmann, S. et al. Role of Drosophila IKKγ in a toll-independent antibacterial immune response. Nat. Immunol. 1, 342–347 (2000).
Lu, Y., Wu, L.P. & Anderson, K.V. The antibacterial arm of the Drosophila innate immune response requires an IκB kinase. Genes Dev. 15, 104–110 (2001).
Deng, L. et al. Activation of the IκB kinase complex by TRAF6 requires a dimeric ubiquitin-conjugating enzyme complex and a unique polyubiquitin chain. Cell 103, 351–361 (2000).
Wang, C. et al. TAK1 is a ubiquitin-dependent kinase of MKK and IKK. Nature 412, 346–351 (2001).
Boutros, M. et al. Genome-wide RNAi analysis of growth and viability in Drosophila cells. Science 303, 832–835 (2004).
Crosby, M.A., Goodman, J.L., Strelets, V.B., Zhang, P. & Gelbart, W.M. FlyBase: genomes by the dozen. Nucleic Acids Res. 35, D486–D491 (2007).
Reichhart, J.M. et al. Splice-activated UAS hairpin vector gives complete RNAi knockout of single or double target transcripts in Drosophila melanogaster. Genesis 34, 160–164 (2002).
Pena-Rangel, M.T., Rodriguez, I. & Riesgo-Escovar, J.R. A misexpression study examining dorsal thorax formation in Drosophila melanogaster. Genetics 160, 1035–1050 (2002).
DasGupta, R., Kaykas, A., Moon, R.T. & Perrimon, N. Functional genomic analysis of the Wnt-wingless signaling pathway. Science 308, 826–833 (2005).
Blumenthal, A. et al. The Wingless homolog WNT5A and its receptor Frizzled-5 regulate inflammatory responses of human mononuclear cells induced by microbial stimulation. Blood 108, 965–973 (2006).
Gross, I., Georgel, P., Kappler, C., Reichhart, J.M. & Hoffmann, J.A. Drosophila immunity: a comparative analysis of the Rel proteins dorsal and Dif in the induction of the genes encoding diptericin and cecropin. Nucleic Acids Res. 24, 1238–1245 (1996).
Combet, C., Blanchet, C., Geourjon, C. & Deleage, G. NPS@: network protein sequence analysis. Trends Biochem. Sci. 25, 147–150 (2000).
Kumar, S., Tamura, K. & Nei, M. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief. Bioinform. 5, 150–163 (2004).
Leclerc, V. et al. Prophenoloxidase activation is not required for survival to microbial infections in Drosophila. EMBO Rep. 7, 231–235 (2006).
Ligoxygakis, P., Pelte, N., Hoffmann, J.A. & Reichhart, J.M. Activation of Drosophila Toll during fungal infection by a blood serine protease. Science 297, 114–116 (2002).
Acknowledgements
We thank S. Stöven for Relish constructs; J.L. Imler for reporter constructs; S. Kurata for support to A.G. and discussions; M. Shiokawa, Y. Fujiwara, L. Troxler, A. Meunier and R. Walther for technical help; M. Hashimoto for secretarial assistance; and our colleagues for discussions and suggestions. Supported by the Japan Society for the Promotion of Science (A.G.), the Centre National de la Recherche Scientifique, the Ministère de l'Education Nationale de la Recherche et de la Technologie, Special Coordination Funds, the Japanese Ministry of Education, Culture, Sports, Science and Technology, the US National Institutes of Health (AI070167 and AI44220) and the Emmy-Noether Program of the Deutsche Forschungsgemeinschaft.
Author information
Authors and Affiliations
Contributions
A.G., V.G., L.E.C. and D.K. did the drosophila experiments. K.M. and O.T. did the mouse experiments. S.A., M.B., O.T. and J.-M.R. conceived and directed the experiments. A.G., O.T., J.A.H. and J.-M.R. wrote the paper. All authors contributed to manuscript criticism.
Corresponding author
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–6, Table 1 and Methods (PDF 434 kb)
Rights and permissions
About this article
Cite this article
Goto, A., Matsushita, K., Gesellchen, V. et al. Akirins are highly conserved nuclear proteins required for NF-κB-dependent gene expression in drosophila and mice. Nat Immunol 9, 97–104 (2008). https://doi.org/10.1038/ni1543
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ni1543
This article is cited by
-
Identification of AKIRIN2 as a potential biomarker and correlation with immunotherapy in gastric adenocarcinoma by integrated bioinformatics analysis
Scientific Reports (2022)
-
Additional evidence on the efficacy of different Akirin vaccines assessed on Anopheles arabiensis (Diptera: Culicidae)
Parasites & Vectors (2021)
-
Verloren negatively regulates the expression of IMD pathway dependent antimicrobial peptides in Drosophila
Scientific Reports (2021)
-
AKIRIN2 controls the nuclear import of proteasomes in vertebrates
Nature (2021)
-
Ubiquitin-related processes and innate immunity in C. elegans
Cellular and Molecular Life Sciences (2021)