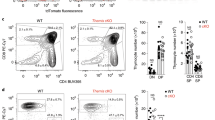
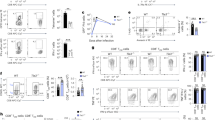
Abstract
T cell immunity requires the long-term survival of T cells that are capable of recognizing self antigens but are not overtly autoreactive. How this balance is achieved remains incompletely understood. Here we identify a homeostatic mechanism that transcriptionally tailors CD8 coreceptor expression in individual CD8+ T cells to the self-specificity of their clonotypic T cell receptor (TCR). 'Coreceptor tuning' results from interplay between cytokine and TCR signals, such that signals from interleukin 7 and other common γ-chain cytokines transcriptionally increase CD8 expression and thereby promote TCR engagement of self ligands, whereas TCR signals impair common γ-chain cytokine signaling and thereby decrease CD8 expression. This dynamic interplay induces individual CD8+ T cells to express CD8 in quantities appropriate for the self-specificity of their TCR, promoting the engagement of self ligands, yet avoiding autoreactivity.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
References
Ernst, B., Lee, D.S., Chang, J.M., Sprent, J. & Surh, C.D. The peptide ligands mediating positive selection in the thymus control T cell survival and homeostatic proliferation in the periphery. Immunity 11, 173–181 (1999).
Kirberg, J., Berns, A. & von Boehmer, H. Peripheral T cell survival requires continual ligation of the T cell receptor to major histocompatibility complex-encoded molecules. J. Exp. Med. 186, 1269–1275 (1997).
Schluns, K.S., Kieper, W.C., Jameson, S.C. & Lefrancois, L. Interleukin-7 mediates the homeostasis of naive and memory CD8 T cells in vivo. Nat. Immunol. 1, 426–432 (2000).
Surh, C.D. & Sprent, J. Regulation of mature T cell homeostasis. Semin. Immunol. 17, 183–191 (2005).
Tan, J.T. et al. IL-7 is critical for homeostatic proliferation and survival of naive T cells. Proc. Natl. Acad. Sci. USA 98, 8732–8737 (2001).
Tanchot, C., Lemonnier, F.A., Perarnau, B., Freitas, A.A. & Rocha, B. Differential requirements for survival and proliferation of CD8 naive or memory T cells. Science 276, 2057–2062 (1997).
Jiang, Q. et al. Cell biology of IL-7, a key lymphotrophin. Cytokine Growth Factor Rev. 16, 513–533 (2005).
Park, J.H. et al. Suppression of IL7Rα transcription by IL-7 and other prosurvival cytokines: a novel mechanism for maximizing IL-7-dependent T cell survival. Immunity 21, 289–302 (2004).
Geiselhart, L.A. et al. IL-7 administration alters the CD4:CD8 ratio, increases T cell numbers, and increases T cell function in the absence of activation. J. Immunol. 166, 3019–3027 (2001).
Goldrath, A.W. & Bevan, M.J. Low-affinity ligands for the TCR drive proliferation of mature CD8+ T cells in lymphopenic hosts. Immunity 11, 183–190 (1999).
Kieper, W.C., Burghardt, J.T. & Surh, C.D. A role for TCR affinity in regulating naive T cell homeostasis. J. Immunol. 172, 40–44 (2004).
Marquez, M.E. et al. CD8 T cell sensory adaptation dependent on TCR avidity for self-antigens. J. Immunol. 175, 7388–7397 (2005).
Holler, P.D. & Kranz, D.M. Quantitative analysis of the contribution of TCR/pepMHC affinity and CD8 to T cell activation. Immunity 18, 255–264 (2003).
Maile, R. et al. Peripheral “CD8 tuning” dynamically modulates the size and responsiveness of an antigen-specific T cell pool in vivo. J. Immunol. 174, 619–627 (2005).
Sarafova, S.D. et al. Modulation of coreceptor transcription during positive selection dictates lineage fate independently of TCR/coreceptor specificity. Immunity 23, 75–87 (2005).
Teh, H.S., Kishi, H., Scott, B. & Von Boehmer, H. Deletion of autospecific T cells in T cell receptor (TCR) transgenic mice spares cells with normal TCR levels and low levels of CD8 molecules. J. Exp. Med. 169, 795–806 (1989).
Robey, E.A. et al. A self-reactive T cell population that is not subject to negative selection. Int. Immunol. 4, 969–974 (1992).
Rathmell, J.C., Farkash, E.A., Gao, W. & Thompson, C.B. IL-7 enhances the survival and maintains the size of naive T cells. J. Immunol. 167, 6869–6876 (2001).
Teague, T.K., Marrack, P., Kappler, J.W. & Vella, A.T. IL-6 rescues resting mouse T cells from apoptosis. J. Immunol. 158, 5791–5796 (1997).
Yu, Q., Erman, B., Bhandoola, A., Sharrow, S.O. & Singer, A. In vitro evidence that cytokine receptor signals are required for differentiation of double positive thymocytes into functionally mature CD8+ T cells. J. Exp. Med. 197, 475–487 (2003).
von Freeden-Jeffry, U. et al. Lymphopenia in interleukin (IL)-7 gene-deleted mice identifies IL-7 as a nonredundant cytokine. J. Exp. Med. 181, 1519–1526 (1995).
Gorman, S.D., Sun, Y.H., Zamoyska, R. & Parnes, J.R. Molecular linkage of the Ly-3 and Ly-2 genes. Requirement of Ly-2 for Ly-3 surface expression. J. Immunol. 140, 3646–3653 (1988).
Hennecke, S. & Cosson, P. Role of transmembrane domains in assembly and intracellular transport of the CD8 molecule. J. Biol. Chem. 268, 26607–26612 (1993).
Suzuki, H., Punt, J.A., Granger, L.G. & Singer, A. Asymmetric signaling requirements for thymocyte commitment to the CD4+ versus CD8+ T cell lineages: a new perspective on thymic commitment and selection. Immunity 2, 413–425 (1995).
Ellmeier, W., Sunshine, M.J., Losos, K. & Littman, D.R. Multiple developmental stage-specific enhancers regulate CD8 expression in developing thymocytes and in thymus-independent T cells. Immunity 9, 485–496 (1998).
Hostert, A. et al. A CD8 genomic fragment that directs subset-specific expression of CD8 in transgenic mice. J. Immunol. 158, 4270–4281 (1997).
Hostert, A. et al. Hierarchical interactions of control elements determine CD8alpha gene expression in subsets of thymocytes and peripheral T cells. Immunity 9, 497–508 (1998).
Kioussis, D. & Ellmeier, W. Chromatin and CD4, CD8A and CD8B gene expression during thymic differentiation. Nat. Rev. Immunol. 2, 909–919 (2002).
Zhang, X.L. et al. Distinct stage-specific cis-active transcriptional mechanisms control expression of T cell coreceptor CD8 alpha at double- and single-positive stages of thymic development. J. Immunol. 161, 2254–2266 (1998).
Ellmeier, W., Sunshine, M.J., Losos, K., Hatam, F. & Littman, D.R. An enhancer that directs lineage-specific expression of CD8 in positively selected thymocytes and mature T cells. Immunity 7, 537–547 (1997).
Depper, J.M. et al. Interleukin 2 (IL-2) augments transcription of the IL-2 receptor gene. Proc. Natl. Acad. Sci. USA 82, 4230–4234 (1985).
Nelms, K., Keegan, A.D., Zamorano, J., Ryan, J.J. & Paul, W.E. The IL-4 receptor: signaling mechanisms and biologic functions. Annu. Rev. Immunol. 17, 701–738 (1999).
Cui, Y. et al. Inactivation of Stat5 in mouse mammary epithelium during pregnancy reveals distinct functions in cell proliferation, survival, and differentiation. Mol. Cell. Biol. 24, 8037–8047 (2004).
Kovanen, P.E. & Leonard, W.J. Cytokines and immunodeficiency diseases: critical roles of the γc-dependent cytokines interleukins 2, 4, 7, 9, 15, and 21, and their signaling pathways. Immunol. Rev. 202, 67–83 (2004).
Ehret, G.B. et al. DNA binding specificity of different STAT proteins. Comparison of in vitro specificity with natural target sites. J. Biol. Chem. 276, 6675–6688 (2001).
Teh, H.S. et al. Thymic major histocompatibility complex antigens and the αβ T-cell receptor determine the CD4/CD8 phenotype of T cells. Nature 335, 229–233 (1988).
Lischke, A. et al. The interleukin-4 receptor activates STAT5 by a mechanism that relies upon common γ-chain. J. Biol. Chem. 273, 31222–31229 (1998).
Mamalaki, C. et al. Positive and negative selection in transgenic mice expressing a T-cell receptor specific for influenza nucleoprotein and endogenous superantigen. Dev. Immunol. 3, 159–174 (1993).
Sha, W.C. et al. Positive and negative selection of an antigen receptor on T cells in transgenic mice. Nature 336, 73–76 (1988).
Feik, N. et al. Functional and molecular analysis of the double-positive stage-specific CD8 enhancer E8III during thymocyte development. J. Immunol. 174, 1513–1524 (2005).
Erman, B. et al. Coreceptor signal strength regulates positive selection but does not determine CD4/CD8 lineage choice in a physiologic in vivo model. J. Immunol. 177, 6613–6625 (2006).
Mandelbrot, D.A. et al. B7-dependent T-cell costimulation in mice lacking CD28 and CTLA4. J. Clin. Invest. 107, 881–887 (2001).
Tivol, E.A. et al. Loss of CTLA-4 leads to massive lymphoproliferation and fatal multiorgan tissue destruction, revealing a critical negative regulatory role of CTLA-4. Immunity 3, 541–547 (1995).
Tai, X., Van Laethem, F., Sharpe, A.H. & Singer, A. Induction of autoimmune disease in CTLA-4−/− mice depends on a specific CD28 motif that is required for in vivo costimulation. Proc. Natl. Acad. Sci. USA 104, 13756–13761 (2007).
Guimond, M., Fry, T.J. & Mackall, C.L. Cytokine signals in T-cell homeostasis. J. Immunother. 28, 289–294 (2005).
Grossman, Z. & Paul, W.E. Adaptive cellular interactions in the immune system: the tunable activation threshold and the significance of subthreshold responses. Proc. Natl. Acad. Sci. USA 89, 10365–10369 (1992).
Grossman, Z. & Singer, A. Tuning of activation thresholds explains flexibility in the selection and development of T cells in the thymus. Proc. Natl. Acad. Sci. USA 93, 14747–14752 (1996).
Bosselut, R., Guinter, T.I., Sharrow, S.O. & Singer, A. Unraveling a revealing paradox: Why major histocompatibility complex I-signaled thymocytes “paradoxically” appear as CD4+8lo transitional cells during positive selection of CD8+ T cells. J. Exp. Med. 197, 1709–1719 (2003).
Kaldjian, E. et al. Nonequivalent effects of PKC activation by PMA on murine CD4 and CD8 cell-surface expression. FASEB J. 2, 2801–2806 (1988).
McCarthy, S.A., Kaldjian, E. & Singer, A. Induction of anti-CD8 resistant cytotoxic T lymphocytes by anti-CD8 antibodies. Functional evidence for T cell signaling induced by multi-valent cross-linking of CD8 on precursor cells. J. Immunol. 141, 3737–3746 (1988).
Wiest, D.L. et al. Regulation of T cell receptor expression in immature CD4+CD8+ thymocytes by p56lck tyrosine kinase: basis for differential signaling by CD4 and CD8 in immature thymocytes expressing both coreceptor molecules. J. Exp. Med. 178, 1701–1712 (1993).
Bilic, I. et al. Negative regulation of CD8 expression via Cd8 enhancer-mediated recruitment of the zinc finger protein MAZR. Nat. Immunol. 7, 392–400 (2006).
Hostert, A. et al. A region in the CD8 gene locus that directs expression to the mature CD8 T cell subset in transgenic mice. Immunity 7, 525–536 (1997).
Noguchi, M. et al. Functional cleavage of the common cytokine receptor γ chain (γc) by calpain. Proc. Natl. Acad. Sci. USA 94, 11534–11539 (1997).
Zhu, J. et al. Transient inhibition of interleukin 4 signaling by T cell receptor ligation. J. Exp. Med. 192, 1125–1134 (2000).
Xue, H.H. et al. IL-2 negatively regulates IL-7 receptor α chain expression in activated T lymphocytes. Proc. Natl. Acad. Sci. USA 99, 13759–13764 (2002).
Brugnera, E. et al. Coreceptor reversal in the thymus: signaled CD4+8+ thymocytes initially terminate CD8 transcription even when differentiating into CD8+ T cells. Immunity 13, 59–71 (2000).
Singer, A. New perspectives on a developmental dilemma: the kinetic signaling model and the importance of signal duration for the CD4/CD8 lineage decision. Curr. Opin. Immunol. 14, 207–215 (2002).
Singer, A. & Bosselut, R. CD4/CD8 coreceptors in thymocyte development, selection, and lineage commitment: analysis of the CD4/CD8 lineage decision. Adv. Immunol. 83, 91–131 (2004).
Bosselut, R., Feigenbaum, L., Sharrow, S.O. & Singer, A. Strength of signaling by CD4 and CD8 coreceptor tails determines the number but not the lineage direction of positively selected thymocytes. Immunity 14, 483–494 (2001).
Acknowledgements
We thank R. Gress, R. Hodes, S. Sharrow, N. Taylor, and T. Waldmann for critically reading the manuscript; S. Sharrow, T. Adams and L. Granger for flow cytometry; and Z. Grossman for discussions. Il7−/− mice were provided by the DNAX Research Institute, HY Rag2−/− mice were provided by M. Vacchio (National Institutes of Health), and Ctla4−/−Cd28−/− and Ctla4−/−Cd28+/− mice were provided by A. Sharpe (Harvard Medical School). Supported by the Intramural Research Program of the National Institutes of Health, National Cancer Institute, Center for Cancer Research.
Author information
Authors and Affiliations
Contributions
J.-H.P. conceptualized the research, did experiments, analyzed data and contributed to the writing of the manuscript; S.A., P.J.L, S.D.S. and L.L.D. did experiments and analyzed data; A.S.A. and B.E. constructed some of the experimental mice; X.L. and R.B. provided data; W.E. provided materials; L.F. generated transgenic mice; and A.S. conceptualized the research, directed the study, analyzed data and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–4 (PDF 523 kb)
Rights and permissions
About this article
Cite this article
Park, JH., Adoro, S., Lucas, P. et al. 'Coreceptor tuning': cytokine signals transcriptionally tailor CD8 coreceptor expression to the self-specificity of the TCR. Nat Immunol 8, 1049–1059 (2007). https://doi.org/10.1038/ni1512
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ni1512
This article is cited by
-
T cell receptor and cytokine signal integration in CD8+ T cells is mediated by the protein Themis
Nature Immunology (2020)
-
Regulatory mechanisms in T cell receptor signalling
Nature Reviews Immunology (2018)
-
The common γ-chain cytokine receptor: tricks-and-treats for T cells
Cellular and Molecular Life Sciences (2016)
-
The TCR's sensitivity to self peptide–MHC dictates the ability of naive CD8+ T cells to respond to foreign antigens
Nature Immunology (2015)
-
Epigenetic plasticity of Cd8a locus during CD8+ T-cell development and effector differentiation and reprogramming
Nature Communications (2014)