Abstract
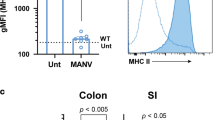
Toll-like receptors (TLRs) recognize distinct microbial components and induce innate immune responses. TLR5 is triggered by bacterial flagellin. Here we generated Tlr5−/− 1mice and assessed TLR5 function in vivo. Unlike other TLRs, TLR5 was not expressed on conventional dendritic cells or macrophages. In contrast, TLR5 was expressed mainly on intestinal CD11c+ lamina propria cells (LPCs). CD11c+ LPCs detected pathogenic bacteria and secreted proinflammatory cytokines in a TLR5-dependent way. However, CD11c+ LPCs do not express TLR4 and did not secrete proinflammatory cytokines after exposure to a commensal bacterium. Notably, transport of pathogenic Salmonella typhimurium from the intestinal tract to mesenteric lymph nodes was impaired in Tlr5−/− mice. These data suggest that CD11c+ LPCs, via TLR5, detect and are used by pathogenic bacteria in the intestinal lumen.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Accession codes
References
Akira, S., Uematsu, S. & Takeuchi, O. Pathogen recognition and innate immunity. Cell 124, 783–801 (2006).
Hayashi, F. et al. The innate immune response to bacterial flagellin is mediated by Toll-like receptor 5. Nature 410, 1099–1103 (2001).
Macnab, R.M. Genetics and biogenesis of bacterial flagella. Annu. Rev. Genet. 26, 131–158 (1992).
Gewirtz, A.T., Navas, T.A., Lyons, S., Godowski, P.J. & Madara, J.L. Cutting edge: bacterial flagellin activates basolaterally expressed TLR5 to induce epithelial proinflammatory gene expression. J. Immunol. 167, 1882–1885 (2001).
Salazar-Gonzalez, R.M. & McSorley, S.J. Salmonella flagellin, a microbial target of the innate and adaptive immune system. Immunol. Lett. 101, 117–122 (2005).
Sierro, F. et al. Flagellin stimulation of intestinal epithelial cells triggers CCL20-mediated migration of dendritic cells. Proc. Natl. Acad. Sci. USA 98, 13722–13727 (2001).
Sebastiani, G. et al. Cloning and characterization of the murine toll-like receptor 5 (Tlr5) gene: sequence and mRNA expression studies in Salmonella-susceptible MOLF/Ei mice. Genomics 64, 230–240 (2000).
Hawn, T.R. et al. A common dominant TLR5 stop codon polymorphism abolishes flagellin signaling and is associated with susceptibility to legionnaires' disease. J. Exp. Med. 198, 1563–1572 (2003).
Jang, M.H. et al. CCR7 is critically important for migration of dendritic cells in intestinal lamina propria to mesenteric lymph nodes. J. Immunol. 176, 803–810 (2006).
Gewirtz, A.T. et al. Salmonella typhimurium translocates flagellin across intestinal epithelia, inducing a proinflammatory response. J. Clin. Invest. 107, 99–109 (2001).
Pavli, P., Woodhams, C.E., Doe, W.F. & Hume, D.A. Isolation and characterization of antigen-presenting dendritic cells from the mouse intestinal lamina propria. Immunology 70, 40–47 (1990).
Niedergang, F., Didierlaurent, A., Kraehenbuhl, J.P. & Sirard, J.C. Dendritic cells: the host Achille's heel for mucosal pathogens? Trends Microbiol. 12, 79–88 (2004).
Ruedl, C., Rieser, C., Bock, G., Wick, G. & Wolf, H. Phenotypic and functional characterization of CD11c+ dendritic cell population in mouse Peyer's patches. Eur. J. Immunol. 26, 1801–1806 (1996).
Wakkach, A. et al. Characterization of dendritic cells that induce tolerance and T regulatory 1 cell differentiation in vivo. Immunity 18, 605–617 (2003).
Kutsukake, K., Ohya, Y., Yamaguchi, S. & Iino, T. Operon structure of flagellar genes in Salmonella typhimurium. Mol. Gen. Genet. 214, 11–15 (1988).
Mayrhofer, G., Pugh, C.W. & Barclay, A.N. The distribution, ontogeny and origin in the rat of Ia-positive cells with dendritic morphology and of Ia antigen in epithelia, with special reference to the intestine. Eur. J. Immunol. 13, 112–122 (1983).
Mowat, A.M. Anatomical basis of tolerance and immunity to intestinal antigens. Nat. Rev. Immunol. 3, 331–341 (2003).
Chirdo, F.G., Millington, O.R., Beacock-Sharp, H. & Mowat, A.M. Immunomodulatory dendritic cells in intestinal lamina propria. Eur. J. Immunol. 35, 1831–1840 (2005).
Worbs, T. et al. Oral tolerance originates in the intestinal immune system and relies on antigen carriage by dendritic cells. J. Exp. Med. 203, 519–527 (2006).
Niess, J.H. et al. CX3CR1-mediated dendritic cell access to the intestinal lumen and bacterial clearance. Science 307, 254–258 (2005).
Andersen-Nissen, E. et al. Evasion of Toll-like receptor 5 by flagellated bacteria. Proc. Natl. Acad. Sci. USA 102, 9247–9252 (2005).
Hopkins, S.A., Niedergang, F., Corthesy-Theulaz, I.E. & Kraehenbuhl, J.P. A recombinant Salmonella typhimurium vaccine strain is taken up and survives within murine Peyer's patch dendritic cells. Cell. Microbiol. 2, 59–68 (2000).
Rescigno, M. et al. Dendritic cells express tight junction proteins and penetrate gut epithelial monolayers to sample bacteria. Nat. Immunol. 2, 361–367 (2001).
Patel, J.C., Rossanese, O.W. & Galan, J.E. The functional interface between Salmonella and its host cell: opportunities for therapeutic intervention. Trends Pharmacol. Sci. 26, 564–570 (2005).
Vazquez-Torres, A. et al. Extraintestinal dissemination of Salmonella by CD18-expressing phagocytes. Nature 401, 804–808 (1999).
Hoshino, K. et al. Cutting edge: Toll-like receptor 4 (TLR4)-deficient mice are hyporesponsive to lipopolysaccharide: evidence for TLR4 as the Lps gene product. J. Immunol. 162, 3749–3752 (1999).
Gulig, P.A. & Curtiss, R., III Plasmid-associated virulence of Salmonella typhimurium. Infect. Immun. 55, 2891–2901 (1987).
Hemmi, H., Kaisho, T., Takeda, K. & Akira, S. The roles of Toll-like receptor 9, MyD88, and DNA-dependent protein kinase catalytic subunit in the effects of two distinct CpG DNAs on dendritic cell subsets. J. Immunol. 170, 3059–3064 (2003).
Acknowledgements
We thank K. Smith and T. Hawn (Institute for Systems Biology, Seattle, Washington) for providing purified flagellin; C. Sasagawa and T. Suzuki (Institute of Medical Science, Tokyo, Japan) for providing bacteria; members of the DNA-chip Development Center for Infectious Diseases (RIMD, Osaka University, Osaka, Japan) for technical advice; N. Kitagaki for technical assistance; and M. Hashimoto for secretarial assistance. Supported by Special Coordination Funds, the Ministry of Education, Culture, Sports, Science and Technology, and Research Fellowships of the Japan Society for the Promotion of Science for Young Scientists.
Author information
Authors and Affiliations
Contributions
S.U. and M.H.J. did most of the experiments to characterize mouse phenotypes; N.C. helped with the quantitative PCR, microarray analysis, isolation of cells and enzyme-linked immunosorbent assays; Z.G. helped to isolate cells and with immunostaining and did the surgical operations for the intestinal loop assay; Y.K. helped with analysis of microarray data; M.Y. helped to generate Tlr5−/− mice; H.K. helped with the enzyme-linked immunosorbent assays; N.S. helped to isolate cells; H.M. provided S. typhimurium and provided instructions for infection experiments; H.K. helped with the infection experiments; H.H. helped to generate Tlr5−/− mice; C.C. helped with the infection experiments; T.K., K.J.I. and O.T. provided advice for the experiments; M.M. provided advice for the experiments and manuscript; K.T. helped to generate Tlr5−/− mice and to design experiments; and S.A. designed all the experiments and prepared the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Fig. 1
Generation of Tlr5−/− mice. (PDF 1622 kb)
Supplementary Fig. 2
CD11c+ LPCs produce IL-6 in response to TLR2 and TLR9 stimulation. (PDF 791 kb)
Supplementary Fig. 3
Surface phenotype of MLN cells 2 d after oral S. typhimurium infection. (PDF 1128 kb)
Supplementary Fig. 4
Uptake of S. typhimurium in situ. (PDF 903 kb)
Supplementary Table 1
Primer sequences. (PDF 21 kb)
Rights and permissions
About this article
Cite this article
Uematsu, S., Jang, M., Chevrier, N. et al. Detection of pathogenic intestinal bacteria by Toll-like receptor 5 on intestinal CD11c+ lamina propria cells. Nat Immunol 7, 868–874 (2006). https://doi.org/10.1038/ni1362
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ni1362
This article is cited by
-
TLR5 agonist in combination with anti-PD-1 treatment enhances anti-tumor effect through M1/M2 macrophage polarization shift and CD8+ T cell priming
Cancer Immunology, Immunotherapy (2024)
-
Aryl hydrocarbon receptor utilises cellular zinc signals to maintain the gut epithelial barrier
Nature Communications (2023)
-
Group 3 innate lymphoid cell pyroptosis represents a host defence mechanism against Salmonella infection
Nature Microbiology (2022)
-
Activation of Toll-like receptor 5 in microglia modulates their function and triggers neuronal injury
Acta Neuropathologica Communications (2020)
-
Toll-like receptors: exploring their potential connection with post-operative infectious complications and cancer recurrence
Clinical & Experimental Metastasis (2020)