Abstract
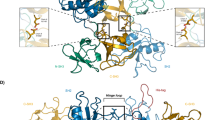
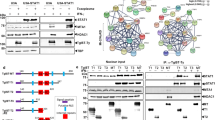
The N-terminal protein interaction domain (N-domain) of the signal transducer and activator of transcription-4 (STAT4) is believed to stabilize interactions between two phosphorylated STAT4 dimers to form STAT4 tetramers. Here, we show that nonphosphorylated STAT4 dimers form in vivo before cytokine receptor–driven activation. Mutations in the N-domain dimerization interface abolished assembly of nonphosphorylated STAT4 dimers and prevented STAT4 phosphorylation mediated by cytokine receptors. In addition, N-domain dimerization occurred for other STAT family members but was homotypic in character. This implies a conserved role for N-domain dimerization, which might include influencing interactions with cytokine receptors, favoring homodimer formation or accelerating formation of the phosphorylated STAT dimer.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Accession codes
References
Jacobson, N.G. et al. Interleukin 12 signaling in T helper type 1 (Th1) cells involves tyrosine phosphorylation of signal transducer and activator of transcription (Stat)3 and Stat4. J. Exp. Med. 181, 1755–1762 (1995).
Bacon, C.M. et al. Interleukin 12 induces tyrosine phosphorylation and activation of STAT4 in human lymphocytes. Proc. Natl. Acad. Sci. USA 92, 7307–7311 (1995).
Kaplan, M.H. et al. Impaired IL-12 responses and enhanced development of Th2 cells in Stat4-deficient mice. Nature 382, 174–177 (1996).
Horvath, C.M., Wen, Z. & Darnell, J.E. Jr. A STAT protein domain that determines DNA sequence recognition suggests a novel DNA-binding domain. Genes Dev. 9, 984–994 (1995).
Seidel, H.M. et al. Spacing of palindromic half sites as a determinant of selective STAT (signal transducers and activators of transcription) DNA binding and transcriptional activity. Proc. Natl. Acad. Sci. USA 92, 3041–3045 (1995).
Ihle, J.N. STATs: signal transducers and activators of transcription. Cell 84, 331–334 (1996).
Xu, X., Sun, Y.L. & Hoey, T. Cooperative DNA binding and sequence-selective recognition conferred by the STAT amino-terminal domain. Science 273, 794–797 (1996).
Symes, A. et al. STAT proteins participate in the regulation of the vasoactive intestinal peptide gene by the ciliary neurotrophic factor family of cytokines. Mol. Endocrinol. 8, 1750–1763 (1994).
Dajee, M. et al. Prolactin induction of the α2-Macroglobulin gene in rat ovarian granulosa cells: stat 5 activation and binding to the interleukin-6 response element. Mol. Endocrinol. 10, 171–184 (1996).
Chatterjee-Kishore, M. et al. How Stat1 mediates constitutive gene expression: a complex of unphosphorylated Stat1 and IRF1 supports transcription of the LMP2 gene. EMBO J. 19, 4111–4122 (2000).
Meyer, T., Gavenis, K. & Vinkemeier, U. Cell type-specific and tyrosine phosphorylation-independent nuclear presence of STAT1 and STAT3. Exp. Cell Res. 272, 45–55 (2002).
Darnell, J.E. Jr. STATs and gene regulation. Science 277, 1630–1635 (1997).
Leaman, D.W. et al. Regulation of STAT-dependent pathways by growth factors and cytokines. FASEB J. 10, 1578–1588 (1996).
Schindler, C. & Darnell, J.E. Jr. Transcriptional responses to polypeptide ligands: the JAK-STAT pathway. Annu. Rev. Biochem. 64, 621–651 (1995).
Kisseleva, T. et al. Signaling through the JAK/STAT pathway, recent advances and future challenges. Gene 285, 1–24 (2002).
Ndubuisi, M.I. et al. Cellular physiology of STAT3: where's the cytoplasmic monomer? J. Biol. Chem. 274, 25499–25509 (1999).
Haan, S. et al. Cytoplasmic STAT proteins associate prior to activation. Biochem. J. 345, 417–421 (2000).
Shah, M. et al. Interactions of STAT3 with caveolin-1 and heat shock protein 90 in plasma membrane raft and cytosolic complexes. Preservation of cytokine signaling during fever. J. Biol. Chem. 277, 45662–45669 (2002).
Guo, G.G. et al. Association of the chaperone glucose-regulated protein 58 (GRP58/ER-60/ERp57) with Stat3 in cytosol and plasma membrane complexes. J. Interferon Cytokine Res. 22, 555–563 (2002).
Braunstein, J. et al. STATs dimerize in the absence of phosphorylation. J. Biol. Chem. (2003).
Chen, X. et al. Crystal structure of a tyrosine phosphorylated STAT-1 dimer bound to DNA. Cell 93, 827–839 (1998).
Vinkemeier, U. et al. Structure of the amino-terminal protein interaction domain of STAT-4. Science 279, 1048–1052 (1998).
Murphy, T.L. et al. Role of the Stat4 N domain in receptor proximal tyrosine phosphorylation. Mol. Cell Biol. 20, 7121–7131 (2000).
Chang, H.C. et al. Stat4 requires the N-terminal domain for efficient phosphorylation. J. Biol. Chem. (2003).
Chen, X. et al. A reinterpretation of the dimerization interface of the N-terminal domains of STATs. Protein Sci. 12, 361–365 (2003).
Jones, S. & Thornton, J.M. Principles of protein-protein interactions. Proc. Natl. Acad. Sci. USA 93, 13–20 (1996).
Lawrence, M.C. & Colman, P.M. Shape complementarity at protein/protein interfaces. J. Mol. Biol. 234, 946–950 (1993).
Lo Conte, L., Chothia, C. & Janin, J. The atomic structure of protein-protein recognition sites. J. Mol. Biol. 285, 2177–2198 (1999).
Beranger, F. et al. Getting more from the two-hybrid system: N-terminal fusions to LexA are efficient and sensitive baits for two-hybrid studies. Nucleic Acids Res. 25, 2035–2036 (1997).
Farrar, J.D. et al. Selective loss of type I interferon–induced STAT4 activation caused by a minisatellite insertion in mouse STAT2. Nat. Immunol. 1, 65–69 (2000).
Farrar, J.D. & Murphy, K.M. Type I interferons and T helper development. Immunol. Today 21, 484–489 (2000).
Yang, J. et al. Induction of interferon-γ production in Th1 CD4+ T cells: evidence for two distinct pathways for promoter activation. Eur. J. Immunol. 29, 548–555 (1999).
Yang, J. et al. IL-18-stimulated GADD45β required in cytokine-induced, but not TCR-induced, IFN-γ production. Nat. Immunol. 2, 157–164 (2001).
Afkarian, M. et al. T-bet is a STAT1-induced regulator of IL-12R expression in naive CD4+ T cells. Nat. Immunol. 3, 549–557 (2002).
Meraz, M.A. et al. Targeted disruption of the Stat1 gene in mice reveals unexpected physiologic specificity in the JAK-STAT signaling pathway. Cell 84, 431–442 (1996).
John, S. et al. The significance of tetramerization in promoter recruitment by Stat5. Mol. Cell Biol. 19, 1910–1918 (1999).
Soldaini, E. et al. DNA binding site selection of dimeric and tetrameric Stat5 proteins reveals a large repertoire of divergent tetrameric Stat5a binding sites. Mol. Cell Biol. 20, 389–401 (2000).
Farrar, J.D. et al. Recruitment of Stat4 to the human interferon-α/β receptor requires activated Stat2. J. Biol. Chem. 275, 2693–2697 (2000).
Ihle, J.N. & Kerr, I.M. Jaks and Stats in signaling by the cytokine receptor superfamily. Trends Genet. 11, 69–74 (1995).
Cho, S.S. et al. Activation of STAT4 by IL-12 and IFN-α: evidence for the involvement of ligand-induced tyrosine and serine phosphorylation. J. Immunol. 157, 4781–4789 (1996).
Hubbard, S.J., Campbell, S.F. & Thornton, J.M. Molecular recognition. Conformational analysis of limited proteolytic sites and serine proteinase protein inhibitors. J. Mol. Biol. 220, 507–530 (1991).
Evan, G.I. et al. Isolation of monoclonal antibodies specific for human c-myc proto-oncogene product. Mol. Cell Biol. 5, 3610–3616 (1985).
Ranganath, S. et al. GATA-3-dependent enhancer activity in IL-4 gene regulation. J. Immunol. 161, 3822–3826 (1998).
Ouyang, W. et al. The Ets transcription factor ERM is Th1-specific and induced by IL-12 through a Stat4-dependent pathway. Proc. Natl. Acad. Sci. USA 96, 3888–3893 (1999).
Morinobu, A. et al. STAT4 serine phosphorylation is critical for IL-12-induced IFN-γ production but not for cell proliferation. Proc. Natl. Acad. Sci. USA 99, 12281–12286 (2002).
Ouyang, W. et al. Inhibition of Th1 development mediated by GATA-3 through an IL-4-independent mechanism. Immunity 9, 745–755 (1998).
Acknowledgements
The authors thank G.R. Stark (Lerner Research Institute, Cleveland, Ohio, USA) for providing the U3A cells, and J.D. Farrar (University of Texas Southwestern Medical Center, Dallas, Texas, USA) for helpful discussions.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Ota, N., Brett, T., Murphy, T. et al. N-domain–dependent nonphosphorylated STAT4 dimers required for cytokine-driven activation. Nat Immunol 5, 208–215 (2004). https://doi.org/10.1038/ni1032
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ni1032
This article is cited by
-
A novel interface between the N-terminal and coiled-coil domain of STAT1 functions in an auto-inhibitory manner
Cell Communication and Signaling (2023)
-
Turning transcription on or off with STAT5: when more is less
Nature Immunology (2011)
-
Preassembly and ligand-induced restructuring of the chains of the IFN-γ receptor complex: the roles of Jak kinases, Stat1 and the receptor chains
Cell Research (2006)
-
IL-6 signaling via the STAT3/SOCS3 pathway: Functional Analysis of the Conserved STAT3 N-domain
Molecular and Cellular Biochemistry (2006)