Abstract
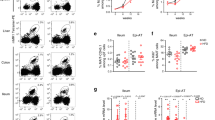
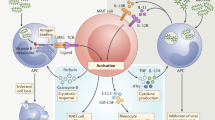
Type 1 diabetes (T1D) is an autoimmune disease that results from the destruction of pancreatic β-cells by the immune system that involves innate and adaptive immune cells. Mucosal-associated invariant T cells (MAIT cells) are innate-like T-cells that recognize derivatives of precursors of bacterial riboflavin presented by the major histocompatibility complex (MHC) class I–related molecule MR1. Since T1D is associated with modification of the gut microbiota, we investigated MAIT cells in this pathology. In patients with T1D and mice of the non-obese diabetic (NOD) strain, we detected alterations in MAIT cells, including increased production of granzyme B, which occurred before the onset of diabetes. Analysis of NOD mice that were deficient in MR1, and therefore lacked MAIT cells, revealed a loss of gut integrity and increased anti-islet responses associated with exacerbated diabetes. Together our data highlight the role of MAIT cells in the maintenance of gut integrity and the control of anti-islet autoimmune responses. Monitoring of MAIT cells might represent a new biomarker of T1D, while manipulation of these cells might open new therapeutic strategies.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
References
Atkinson, M.A., Eisenbarth, G.S. & Michels, A.W. Type 1 diabetes. Lancet 383, 69–82 (2014).
Diana, J. et al. Crosstalk between neutrophils, B-1a cells and plasmacytoid dendritic cells initiates autoimmune diabetes. Nat. Med. 19, 65–73 (2013).
Anderson, M.S. & Bluestone, J.A. The NOD mouse: a model of immune dysregulation. Annu. Rev. Immunol. 23, 447–485 (2005).
Lehuen, A., Diana, J., Zaccone, P. & Cooke, A. Immune cell crosstalk in type 1 diabetes. Nat. Rev. Immunol. 10, 501–513 (2010).
Bluestone, J.A., Herold, K. & Eisenbarth, G. Genetics, pathogenesis and clinical interventions in type 1 diabetes. Nature 464, 1293–1300 (2010).
Wen, L. et al. Innate immunity and intestinal microbiota in the development of Type 1 diabetes. Nature 455, 1109–1113 (2008).
Markle, J.G.M. et al. Sex differences in the gut microbiome drive hormone-dependent regulation of autoimmunity. Science 339, 1084–1088 (2013).
Yurkovetskiy, L. et al. Gender bias in autoimmunity is influenced by microbiota. Immunity 39, 400–412 (2013).
Kostic, A.D. et al. The dynamics of the human infant gut microbiome in development and in progression toward type 1 diabetes. Cell Host Microbe 17, 260–273 (2015).
Alkanani, A.K. et al. Alterations in intestinal microbiota correlate with susceptibility to type 1 diabetes. Diabetes 64, 3510–3520 (2015).
Vatanen, T. et al. Variation in microbiome LPS immunogenicity contributes to autoimmunity in humans. Cell 165, 1551 (2016).
Alam, C. et al. Inflammatory tendencies and overproduction of IL-17 in the colon of young NOD mice are counteracted with diet change. Diabetes 59, 2237–2246 (2010).
Alam, C. et al. Effects of a germ-free environment on gut immune regulation and diabetes progression in non-obese diabetic (NOD) mice. Diabetologia 54, 1398–1406 (2011).
Bosi, E. et al. Increased intestinal permeability precedes clinical onset of type 1 diabetes. Diabetologia 49, 2824–2827 (2006).
Sapone, A. et al. Zonulin upregulation is associated with increased gut permeability in subjects with type 1 diabetes and their relatives. Diabetes 55, 1443–1449 (2006).
Badami, E. et al. Defective differentiation of regulatory FoxP3+ T cells by small-intestinal dendritic cells in patients with type 1 diabetes. Diabetes 60, 2120–2124 (2011).
Treiner, E. et al. Selection of evolutionarily conserved mucosal-associated invariant T cells by MR1. Nature 422, 164–169 (2003).
Kjer-Nielsen, L. et al. MR1 presents microbial vitamin B metabolites to MAIT cells. Nature 491, 717–723 (2012).
Corbett, A.J. et al. T-cell activation by transitory neo-antigens derived from distinct microbial pathways. Nature 509, 361–365 (2014).
Dusseaux, M. et al. Human MAIT cells are xenobiotic-resistant, tissue-targeted, CD161hi IL-17-secreting T cells. Blood 117, 1250–1259 (2011).
Franciszkiewicz, K. et al. MHC class I-related molecule, MR1, and mucosal-associated invariant T cells. Immunol. Rev. 272, 120–138 (2016).
Magalhaes, I. et al. Mucosal-associated invariant T cell alterations in obese and type 2 diabetic patients. J. Clin. Invest. 125, 1752–1762 (2015).
Illés, Z., Shimamura, M., Newcombe, J., Oka, N. & Yamamura, T. Accumulation of Vα7.2-Jα33 invariant T cells in human autoimmune inflammatory lesions in the nervous system. Int. Immunol. 16, 223–230 (2004).
Serriari, N.-E. et al. Innate mucosal-associated invariant T (MAIT) cells are activated in inflammatory bowel diseases. Clin. Exp. Immunol. 176, 266–274 (2014).
Chen, Z. et al. Mucosal-associated invariant T-cell activation and accumulation after in vivo infection depends on microbial riboflavin synthesis and co-stimulatory signals. Mucosal Immunol. 10.1038/mi.2016.39 (2016).
Le Bourhis, L. et al. MAIT cells detect and efficiently lyse bacterially-infected epithelial cells. PLoS Pathog. 9, e1003681 (2013).
Jeffery, H.C. et al. Biliary epithelium and liver B cells exposed to bacteria activate intrahepatic MAIT cells through MR1. J. Hepatol. 64, 1118–1127 (2016).
Kurioka, A. et al. MAIT cells are licensed through granzyme exchange to kill bacterially sensitized targets. Mucosal Immunol. 8, 429–440 (2015).
Koay, H.-F. et al. A three-stage intrathymic development pathway for the mucosal-associated invariant T cell lineage. Nat. Immunol. 17, 1300–1311 (2016).
Reantragoon, R. et al. Antigen-loaded MR1 tetramers define T cell receptor heterogeneity in mucosal-associated invariant T cells. J. Exp. Med. 210, 2305–2320 (2013).
Leete, P. et al. Differential insulitic profiles determine the extent of β-cell destruction and the age at onset of type 1 diabetes. Diabetes 65, 1362–1369 (2016).
Komulainen, J. et al. Clinical, autoimmune, and genetic characteristics of very young children with type 1 diabetes. Childhood Diabetes in Finland (DiMe) Study Group. Diabetes Care 22, 1950–1955 (1999).
Ravassard, P. et al. A genetically engineered human pancreatic β cell line exhibiting glucose-inducible insulin secretion. J. Clin. Invest. 121, 3589–3597 (2011).
Rahimpour, A. et al. Identification of phenotypically and functionally heterogeneous mouse mucosal-associated invariant T cells using MR1 tetramers. J. Exp. Med. 212, 1095–1108 (2015).
Cui, Y. et al. Mucosal-associated invariant T cell-rich congenic mouse strain allows functional evaluation. J. Clin. Invest. 125, 4171–4185 (2015).
Ivanov, I.I. et al. The orphan nuclear receptor RORγt directs the differentiation program of proinflammatory IL-17+ T helper cells. Cell 126, 1121–1133 (2006).
Dudakov, J.A., Hanash, A.M. & van den Brink, M.R.M. Interleukin-22: immunobiology and pathology. Annu. Rev. Immunol. 33, 747–785 (2015).
Rutz, S., Wang, X. & Ouyang, W. The IL-20 subfamily of cytokines--from host defence to tissue homeostasis. Nat. Rev. Immunol. 14, 783–795 (2014).
Costa, F.R.C. et al. Gut microbiota translocation to the pancreatic lymph nodes triggers NOD2 activation and contributes to T1D onset. J. Exp. Med. 213, 1223–1239 (2016).
Amrani, A. et al. Progression of autoimmune diabetes driven by avidity maturation of a T-cell population. Nature 406, 739–742 (2000).
Diana, J. et al. Viral infection prevents diabetes by inducing regulatory T cells through NKT cell-plasmacytoid dendritic cell interplay. J. Exp. Med. 208, 729–745 (2011).
Turley, S., Poirot, L., Hattori, M., Benoist, C. & Mathis, D. Physiological beta cell death triggers priming of self-reactive T cells by dendritic cells in a type-1 diabetes model. J. Exp. Med. 198, 1527–1537 (2003).
Fujimoto, K. et al. A new subset of CD103+CD8α+ dendritic cells in the small intestine expresses TLR3, TLR7, and TLR9 and induces Th1 response and CTL activity. J. Immunol. 186, 6287–6295 (2011).
Cerovic, V., Bain, C.C., Mowat, A.M. & Milling, S.W.F. Intestinal macrophages and dendritic cells: what's the difference? Trends Immunol. 35, 270–277 (2014).
Murphy, T.L. et al. Transcriptional control of dendritic cell development. Annu. Rev. Immunol. 34, 93–119 (2016).
Cho, Y.-N. et al. Mucosal-associated invariant T cell deficiency in systemic lupus erythematosus. J. Immunol. 193, 3891–3901 (2014).
Magalhaes, I., Kiaf, B. & Lehuen, A. iNKT and MAIT cell alterations in diabetes. Front. Immunol. 6, 341 (2015).
Toubal, A. & Lehuen, A. Lights on MAIT cells, a new immune player in liver diseases. J. Hepatol. 64, 1008–1010 (2016).
Maxwell, J.R. et al. Differential Roles for Interleukin-23 and Interleukin-17 in Intestinal Immunoregulation. Immunity 43, 739–750 (2015).
Lee, J.S. et al. Interleukin-23-independent IL-17 production regulates intestinal epithelial permeability. Immunity 43, 727–738 (2015).
Mahon, J.L. et al. The TrialNet Natural History Study of the Development of Type 1 Diabetes: objectives, design, and initial results. Pediatr. Diabetes 10, 97–104 (2009).
TrialNet - Information for Patients. Available at: https://www.diabetestrialnet.org/PathwayToPrevention/ (2017).
Lochner, M. et al. In vivo equilibrium of proinflammatory IL-17+ and regulatory IL-10+Foxp3+ RORγt+ T cells. J. Exp. Med. 205, 1381–1393 (2008).
Huang, S. et al. MR1 antigen presentation to mucosal-associated invariant T cells was highly conserved in evolution. Proc. Natl. Acad. Sci. USA 106, 8290–8295 (2009).
Pickard, J.M. et al. Rapid fucosylation of intestinal epithelium sustains host-commensal symbiosis in sickness. Nature 514, 638–641 (2014).
Acknowledgements
We thank all the patients and the medical staff of Necker hospital; A. Toubal for critical reading of the manuscript; F. Letourneur and A. Stabilini for technical help; the mouse, Cybio and HistIM facilities of the Cochin Institute; G, Eberl (Institut Pasteur, Paris) for the bacterial artificial chromosome expressing RORγt-GFP; the US National Institutes of Health tetramer core facility; and H. Fohrer-Ting, R. Porcher and M. Laanani for help in statistical analyses. Supported by the Société Francophone de Diabétologie (R.S.), the Foundation Bettencourt Schueller (R.S.), the Laboratoire d'Excellence REVIVE (R.S.), Fondation pour la Recherche Médicale (C.T., M.O. and M.D.), Type 1 Diabetes TrialNet (M.B.), Agence Nationale de la Recherche (NR-11-IDEX-0005-02 Laboratory of Excellence INFLAMEX to A.L.), Fondation pour la Recherche Médicale (DEQ20140329520 to A.L.), the INSERM cross-cutting program on microbiota (A.L.), the European Foundation for the Study of Diabetes–Juvenile Diabetes Research Foundation–Lilly (A.L.), the Département Hospitalo-Universitaire on Autoimmune and Hormonal Diseases (A.L.), the Ministry of Research (O.R.) and Aide aux Jeunes Diabétiques (I.N.).
Author information
Authors and Affiliations
Contributions
O.R., J.D.s. and L.B. performed most of the experiments and data analysis; I.N., C.T., L.C. and B.K. performed experiments and data analysis; M.O. and M.D. participated in experiments with the EndoC-β1 cell line; M.S. and O.L. provided Mr1−/− C57BL/6 and Mr1−/− B6-MAITCAST mice and reagents; A.C., J.R. and J.M. provided mouse MR1 tetramers; J.R., J.M., R.S. and O.L. provided intellectual input; M.B., M.P. and J.B. characterized patients and provided human samples; O.R., I.N. and A.L. wrote the manuscript; and A.L. supervised the work.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–8 and Supplementary Tables 1–4 (PDF 4004 kb)
Rights and permissions
About this article
Cite this article
Rouxel, O., Da silva, J., Beaudoin, L. et al. Cytotoxic and regulatory roles of mucosal-associated invariant T cells in type 1 diabetes. Nat Immunol 18, 1321–1331 (2017). https://doi.org/10.1038/ni.3854
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ni.3854
This article is cited by
-
Mucosal-associated invariant T cells in infectious diseases of respiratory system: recent advancements and applications
Journal of Inflammation (2024)
-
The immunology of type 1 diabetes
Nature Reviews Immunology (2024)
-
The multi-omics analyses of acsl1 reveal its translational significance as a tumor microenvironmental and prognostic biomarker in clear cell renal cell carcinoma
Diagnostic Pathology (2023)
-
Mucosal-associated invariant T cells display both pathogenic and protective roles in patients with inflammatory bowel diseases
Amino Acids (2023)
-
Mucosal-associated invariant T cells contribute to suppression of inflammatory myeloid cells in immune-mediated kidney disease
Nature Communications (2023)