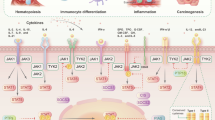
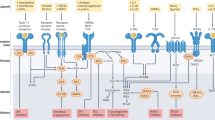
Abstract
Kinases of the Jak ('Janus kinase') family and transcription factors (TFs) of the STAT ('signal transducer and activator of transcription') family constitute a rapid membrane-to-nucleus signaling module that affects every aspect of the mammalian immune system. Research on this paradigmatic pathway has experienced breakneck growth in the quarter century since its discovery and has yielded a stream of basic and clinical insights that have profoundly influenced modern understanding of human health and disease, exemplified by the bench-to-bedside success of Jak inhibitors ('jakinibs') and pathway-targeting drugs. Here we review recent advances in Jak–STAT biology, focusing on immune cell function, disease etiology and therapeutic intervention, as well as broader principles of gene regulation and signal-dependent TFs.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Stark, G.R. & Darnell, J.E. Jr. The JAK-STAT pathway at twenty. Immunity 36, 503–514 (2012).
Darnell, J.E. Jr., Kerr, I.M. & Stark, G.R. Jak-STAT pathways and transcriptional activation in response to IFNs and other extracellular signaling proteins. Science 264, 1415–1421 (1994).
Levy, D.E. & Darnell, J.E. Jr. Signalling: Stats: transcriptional control and biological impact. Nat. Rev. Mol. Cell Biol. 3, 651–662 (2002).
Leonard, W.J. & O'Shea, J.J. JAKS and STATS: biological implications. Annu. Rev. Immunol. 16, 293–322 (1998).
Macchi, P. et al. Mutations of Jak-3 gene in patients with autosomal severe combined immune deficiency (SCID). Nature 377, 65–68 (1995).
Russell, S.M. et al. Mutation of Jak3 in a patient with SCID: essential role of Jak3 in lymphoid development. Science 270, 797–800 (1995).
Luo, H., Hanratty, W.P. & Dearolf, C.R. An amino acid substitution in the Drosophila hopTum-l Jak kinase causes leukemia-like hematopoietic defects. EMBO J. 14, 1412–1420 (1995).
Kralovics, R. et al.A gain-of-function mutation of JAK2 in myeloproliferative disorders. N. Engl. J. Med. 352, 1779–1790 (2005).
James, C. et al. A unique clonal JAK2 mutation leading to constitutive signalling causes polycythaemia vera. Nature 434, 1144–1148 (2005).
Baxter, E.J. et al. Acquired mutation of the tyrosine kinase JAK2 in human myeloproliferative disorders. Lancet 365, 1054–1061 (2005).
Geyer, H.L. & Mesa, R.A. Therapy for myeloproliferative neoplasms: when, which agent, and how? Blood 124, 3529–3537 (2014).
Various. Ruxolitinib clinical trials (US). ClinicalTrials.gov. https://clinicaltrials.gov/ct2/results?term=&recr=&type=&rslt=&age_v=&gndr=&cond=&intr=Ruxolitinib (2016; accessed 2 December 2016).
O'Shea, J.J. et al. The JAK-STAT pathway: impact on human disease and therapeutic intervention. Annu. Rev. Med. 66, 311–328 (2015).
Various. Tofacitinib clinical trials (US). ClinicalTrials.gov. https://clinicaltrials.gov/ct2/results?term=&recr=&type=&rslt=&age_v=&gndr=&cond=&intr=tofacitinib(2016; accessed 21 December 2016).
Various. Jak inhibitor clinical trials (US). ClinicalTrials.gov. https://clinicaltrials.gov/ct2/results?term=JAK1+OR+JAK2+OR+JAK3+OR+TYK2+NOT+Tofacitinib+NOT+Ruxolitinib (2016; accessed 21 December 2016).
Ghoreschi, K., Laurence, A. & O'Shea, J.J. Selectivity and therapeutic inhibition of kinases: to be or not to be? Nat. Immunol. 10, 356–360 (2009).
Tasian, S.K. et al. Potent efficacy of combined PI3K/mTOR and JAK or ABL inhibition in murine xenograft models of Ph-like acute lymphoblastic leukemia. Blood 129, 177–187 (2017).
Gao, S.P. et al. JAK2 inhibition sensitizes resistant EGFR-mutant lung adenocarcinoma to tyrosine kinase inhibitors. Sci. Signal. 9, ra33 (2016).
Britschgi, A. et al. JAK2/STAT5 inhibition circumvents resistance to PI3K/mTOR blockade: a rationale for cotargeting these pathways in metastatic breast cancer. Cancer Cell 22, 796–811 (2012).
Smith, G.A., Uchida, K., Weiss, A. & Taunton, J. essential biphasic role for JaK3 catalytic activity in IL-2 receptor signaling. Nat. Chem. Biol. 12, 373–379 (2016).
Ross, S.H. et al. Phosphoproteomic Analyses of interleukin 2 signaling reveal integrated JAK kinase-dependent and -independent networks in CD8+ T cells. Immunity 45, 685–700 (2016).
Vahedi, G. et al. Super-enhancers delineate disease-associated regulatory nodes in T cells. Nature 520, 558–562 (2015).
Moodley, D. et al. Network pharmacology of JAK inhibitors. Proc. Natl. Acad. Sci. USA 113, 9852–9857 (2016).
Mostafavi, S. et al. Parsing the interferon transcriptional network and its disease associations. Cell 164, 564–578 (2016).
Koppikar, P. et al. Heterodimeric JAK-STAT activation as a mechanism of persistence to JAK2 inhibitor therapy. Nature 489, 155–159 (2012).
Springuel, L. et al. Cooperating JAK1 and JAK3 mutants increase resistance to JAK inhibitors. Blood 124, 3924–3931 (2014).
Benci, J.L. et al. Tumor interferon signaling regulates a multigenic resistance program to immune checkpoint blockade. Cell 167, 1540–1554 (2016).
Spangler, J.B., Moraga, I., Mendoza, J.L. & Garcia, K.C. Insights into cytokine-receptor interactions from cytokine engineering. Annu. Rev. Immunol. 33, 139–167 (2015).
Blouin, C.M. et al. Glycosylation-Dependent IFN-γR partitioning in lipid and actin nanodomains is critical for JAK activation. Cell 166, 920–934 (2016).
Ferrao, R. et al. The structural basis for class II cytokine receptor recognition by JAK1. Structure 24, 897–905 (2016).
Floss, D.M. et al. Defining the functional binding sites of interleukin 12 receptor β1 and interleukin 23 receptor to Janus kinases. Mol. Biol. Cell 27, 2301–2316 (2016).
Lupardus, P.J. et al. Structural snapshots of full-length Jak1, a transmembrane gp130/IL-6/IL-6Rα cytokine receptor complex, and the receptor-Jak1 holocomplex. Structure 19, 45–55 (2011).
Wallweber, H.J.A., Tam, C., Franke, Y., Starovasnik, M.A. & Lupardus, P.J. Structural basis of recognition of interferon-α receptor by tyrosine kinase 2. Nat. Struct. Mol. Biol. 21, 443–448 (2014).
Zhang, D., Wlodawer, A. & Lubkowski, J. Crystal structure of a complex of the intracellular domain of interferon λ Receptor 1 (IFNLR1) and the FERM/SH2 domains of human JAK1. J. Mol. Biol. 428, 4651–4668 (2016).
Brooks, A.J. et al. Mechanism of activation of protein kinase JAK2 by the growth hormone receptor. Science 344, 1249783 (2014).
Ungureanu, D. et al. The pseudokinase domain of JAK2 is a dual-specificity protein kinase that negatively regulates cytokine signaling. Nat. Struct. Mol. Biol. 18, 971–976 (2011).
Bandaranayake, R.M. et al. Crystal structures of the JAK2 pseudokinase domain and the pathogenic mutant V617F. Nat. Struct. Mol. Biol. 19, 754–759 (2012).
Toms, A.V. et al. Structure of a pseudokinase-domain switch that controls oncogenic activation of Jak kinases. Nat. Struct. Mol. Biol. 20, 1221–1223 (2013).
Shan, Y. et al. Molecular basis for pseudokinase-dependent autoinhibition of JAK2 tyrosine kinase. Nat. Struct. Mol. Biol. 21, 579–584 (2014).
Wang, Y. & Levy, D.E. Comparative evolutionary genomics of the STAT family of transcription factors. JAK-STAT 1, 23–33 (2012).
Chen, X. et al. A reinterpretation of the dimerization interface of the N-terminal domains of STATs. Protein Sci. 12, 361–365 (2003).
Mao, X. et al. Structural bases of unphosphorylated STAT1 association and receptor binding. Mol. Cell 17, 761–771 (2005).
Neculai, D. et al. Structure of the unphosphorylated STAT5a dimer. J. Biol. Chem. 280, 40782–40787 (2005).
Li, J. et al. Structural basis for DNA recognition by STAT6. Proc. Natl. Acad. Sci. USA 113, 13015–13020 (2016).
Decker, T., Kovarik, P. & Meinke, A. GAS elements: a few nucleotides with a major impact on cytokine-induced gene expression. J. Interferon Cytokine Res. 17, 121–134 (1997).
Robertson, G. et al. Genome-wide profiles of STAT1 DNA association using chromatin immunoprecipitation and massively parallel sequencing. Nat. Methods 4, 651–657 (2007).
Shlyueva, D., Stampfel, G. & Stark, A. Transcriptional enhancers: from properties to genome-wide predictions. Nat. Rev. Genet. 15, 272–286 (2014).
Vahedi, G. et al. STATs shape the active enhancer landscape of T cell populations. Cell 151, 981–993 (2012).
Ostuni, R. et al. Latent Enhancers activated by stimulation in differentiated cells. Cell 152, 157–171 (2013).
Qiao, Y. et al. Synergistic activation of inflammatory cytokine genes by interferon-gamma-induced chromatin remodeling and Toll-like receptor signaling. Immunity 39, 454–469 (2013).
Wilson, C.B., Rowell, E. & Sekimata, M. Epigenetic control of T-helper-cell differentiation. Nat. Rev. Immunol. 9, 91–105 (2009).
Wei, L. et al. Discrete Roles of STAT4 and SwTAT6 transcription factors in tuning epigenetic modifications and transcription during T helper cell differentiation. Immunity 32, 840–851 (2010).
Durant, L. et al. Diverse targets of the transcription factor STAT3 contribute to T cell pathogenicity and homeostasis. Immunity 32, 605–615 (2010).
Qiao, Y., Kang, K., Giannopoulou, E., Fang, C. & Ivashkiv, L.B. IFN-γ induces histone 3 lysine 27 trimethylation in a small subset of promoters to stably silence gene expression in human macrophages. Cell Rep. 16, 3121–3129 (2016).
Kang, K., Yamaji, D., Yoo, K.H., Robinson, G.W. & Hennighausen, L. Mammary-specific gene activation is defined by progressive recruitment of stat5 during pregnancy and the establishment of H3K4me3 marks. Mol. Cell. Biol. 34, 464–473 (2014).
Willi, M., Yoo, K.H., Wang, C., Trajanoski, Z. & Hennighausen, L. Differential cytokine sensitivities of STAT5-dependent enhancers rely on Stat5 autoregulation. Nucleic Acids Res. 44, 10277–10291 (2016).
Bhattacharya, S. et al. Cooperation of Stat2 and p300/CBP in signalling induced by interferon-α. Nature 383, 344–347 (1996).
Mandal, M. et al. Epigenetic repression of the Igk locus by STAT5-mediated recruitment of the histone methyltransferase Ezh2. Nat. Immunol. 12, 1212–1220 (2011).
Veals, S.A. et al. Subunit of an alpha-interferon-responsive transcription factor is related to interferon regulatory factor and Myb families of DNA-binding proteins. Mol. Cell. Biol. 12, 3315–3324 (1992).
Schaefer, T.S., Sanders, L.K. & Nathans, D. Cooperative transcriptional activity of Jun and Stat3β, a short form of Stat3. Proc. Natl. Acad. Sci. USA 92, 9097–9101 (1995).
Stöcklin, E., Wissler, M., Gouilleux, F. & Groner, B. Functional interactions between Stat5 and the glucocorticoid receptor. Nature 383, 726–728 (1996).
Farlik, M. et al. Nonconventional initiation complex assembly by STAT and NF-kB transcription factors regulates nitric oxide synthase expression. Immunity 33, 25–34 (2010).
Chen, X. et al. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell 133, 1106–1117 (2008).
Ciofani, M. et al. A validated regulatory network for Th17 cell specification. Cell 151, 289–303 (2012).
Li, P. et al. BATF-JUN is critical for IRF4-mediated transcription in T cells. Nature 490, 543–546 (2012).
Garber, M. et al. A high-throughput chromatin immunoprecipitation approach reveals principles of dynamic gene regulation in mammals. Mol. Cell 47, 810–822 (2012).
Langlais, D., Barreiro, L.B. & Gros, P. The macrophage IRF8/IRF1 regulome is required for protection against infections and is associated with chronic inflammation. J. Exp. Med. 213, 585–603 (2016).
Hossain, D.M.S. et al. FoxP3 acts as a cotranscription factor with STAT3 in tumor-induced regulatory T cells. Immunity 39, 1057–1069 (2013).
Kwon, H. et al. analysis of interleukin-21-induced Prdm1 gene regulation reveals functional cooperation of STAT3 and IRF4 transcription factors. Immunity 31, 941–952 (2009).
Xu, X., Sun, Y.L. & Hoey, T. Cooperative DNA binding and sequence-selective recognition conferred by the STAT amino-terminal domain. Science 273, 794–797 (1996).
John, S., Vinkemeier, U., Soldaini, E., Darnell, J.E. Jr. & Leonard, W.J. The significance of tetramerization in promoter recruitment by Stat5. Mol. Cell. Biol. 19, 1910–1918 (1999).
Shin, H.Y. et al. Hierarchy within the mammary STAT5-driven Wap super-enhancer. Nat. Genet. 48, 904–911 (2016).
Kang, K., Robinson, G.W. & Hennighausen, L. Comprehensive meta-analysis of signal transducers and activators of transcription (STAT) genomic binding patterns discerns cell-specific cis-regulatory modules. BMC Genomics 14, 4 (2013).
O'Sullivan, A., Chang, H.C., Yu, Q. & Kaplan, M.H. STAT4 is required for interleukin-12-induced chromatin remodeling of the CD25 locus. J. Biol. Chem. 279, 7339–7345 (2004).
Akaishi, H. et al. Defective IL-2-mediated IL-2 receptor α chain expression in Stat3-deficient T lymphocytes. Int. Immunol. 10, 1747–1751 (1998).
Nakajima, H. et al. An indirect effect of Stat5a in IL-2-induced proliferation: a critical role for Stat5a in IL-2-mediated IL-2 receptor alpha chain induction. Immunity 7, 691–701 (1997).
Villarino, A.V. et al. Helper T cell IL-2 production is limited by negative feedback and STAT-dependent cytokine signals. J. Exp. Med. 204, 65–71 (2007).
Johnston, R.J., Choi, Y.S., Diamond, J.A., Yang, J.A. & Crotty, S. STAT5 is a potent negative regulator of TFH cell differentiation. J. Exp. Med. 209, 243–250 (2012).
Neumann, C. et al. Role of Blimp-1 in programing Th effector cells into IL-10 producers. J. Exp. Med. 211, 1807–1819 (2014).
Poholek, A.C. et al. IL-10 induces a STAT3-dependent autoregulatory loop in TH2 cells that promotes Blimp-1 restriction of cell expansion via antagonism of STAT5 target genes. Science Immunol. http://dx.doi.org/110.1126/sciimmunol.aaf8612 (2016).
Langlais, D., Couture, C., Balsalobre, A. & Drouin, J. The Stat3/GR interaction code: predictive value of direct/indirect DNA recruitment for transcription outcome. Mol. Cell 47, 38–49 (2012).
Russell, B. in Dear Bertrand Russell: A Selection of His Correspondence with the General Public, 1950–1968 (eds. Feinberg, B. Kasrils, D.) 41‐42 Allen & Unwin, London, 1969).
Seidel, H.M. et al. Spacing of palindromic half sites as a determinant of selective STAT (signal transducers and activators of transcription) DNA binding and transcriptional activity. Proc. Nawtl. Acad. Sci. USA 92, 3041–3045 (1995).
Schindler, U., Wu, P., Rothe, M., Brasseur, M. & McKnight, S.L. Components of a Stat recognition code: evidence for two layers of molecular selectivity. Immunity 2, 689–697 (1995).
Costa-Pereira, A.P. et al. Mutational switch of an IL-6 response to an interferon-γ-like response. Proc. Natl. Acad. Sci. USA 99, 8043–8047 (2002).
Qing, Y. & Stark, G.R. Alternative activation of STAT1 and STAT3 in response to interferon-γ. J. Biol. Chem. 279, 41679–41685 (2004).
Park, J.-H. et al. Signaling by intrathymic cytokines, not T cell antigen receptors, specifies CD8 lineage choice and promotes the differentiation of cytotoxic-lineage T cells. Nat. Immunol. 11, 257–264 (2010).
Yang, X.-P. et al. Opposing regulation of the locus encoding IL-17 through direct, reciprocal actions of STAT3 and STAT5. Nat. Immunol. 12, 247–254 (2011).
Zielinski, C.E. et al. Pathogen-induced human TH17 cells produce IFN-(or IL-10 and are regulated by IL-1β. Nature 484, 514–518 (2012).
Wan, C.-K. et al. The cytokines IL-21 and GM-CSF have opposing regulatory roles in the apoptosis of conventional dendritic cells. Immunity 38, 514–527 (2013).
Walker, S.R. et al. STAT5 outcompetes STAT3 To regulate the expression of the oncogenic transcriptional modulator BCL6. Mol. Cell. Biol. 33, 2879–2890 (2013).
Basu, R. et al. IL-1 signaling modulates activation of STAT transcription factors to antagonize retinoic acid signaling and control the TH17 cell-iTreg cell balance. Nat. Immunol. 16, 286–295 (2015).
Hirahara, K. et al. Asymmetric action of STAT transcription factors drives transcriptional outputs and cytokine specificity. Immunity 42, 877–889 (2015).
Jacobson, N.G. et al. Interleukin 12 signaling in T helper type 1 (Th1) cells involves tyrosine phosphorylation of signal transducer and activator of transcription (Stat)3 and Stat4. J. Exp. Med. 181, 1755–1762 (1995).
Lischke, A. et al. The interleukin-4 receptor activates STAT5 by a mechanism that relies upon common gamma-chain. J. Biol. Chem. 273, 31222–31229 (1998).
Nguyen, K.B. et al. Critical role for STAT4 activation by type 1 interferons in the interferon-γ response to viral infection. Science 297, 2063–2066 (2002).
Villarino, A.V. et al. The IL-27R (WSX-1) is required to suppress T cell hyperactivity during infection. Immunity 19, 645–655 (2003).
O'Shea, J.J. & Murray, P.J. Cytokine signaling modules in inflammatory responses. Immunity 28, 477–487 (2008).
Wan, C.-K. et al. Opposing roles of STAT1 and STAT3 in IL-21 function in CD4+ T cells. Proc. Natl. Acad. Sci. USA 112, 9394–9399 (2015).
Shuai, K. & Liu, B. Regulation of JAK-STAT signalling in the immune system. Nat. Rev. Immunol. 3, 900–911 (2003).
Bohmer, F.D. & Friedrich, K. Protein tyrosine phosphatases as wardens of STAT signaling. JAK-STAT 3, e28087 (2014).
Lu, D. et al. The phosphatase DUSP2 controls the activity of the transcription activator STAT3 and regulates TH17 differentiation. Nat. Immunol. 16, 1263–1273 (2015).
Liu, B. et al. PIAS1 selectively inhibits interferon-inducible genes and is important in innate immunity. Nat. Immunol. 5, 891–898 (2004).
Liu, B., Tahk, S., Yee, K.M., Fan, G. & Shuai, K. The ligase PIAS1 restricts natural regulatory T cell differentiation by epigenetic repression. Science 330, 521–525 (2010).
Yoshimura, A., Naka, T. & Kubo, M. SOCS proteins, cytokine signalling and immune regulation. Nat. Rev. Immunol. 7, 454–465 (2007).
Kershaw, N.J. et al. SOCS3 binds specific receptor-JAK complexes to control cytokine signaling by direct kinase inhibition. Nat. Struct. Mol. Biol. 20, 469–476 (2013).
Croker, B.A. et al. SOCS3 negatively regulates IL-6 signaling in vivo. Nat. Immunol. 4, 540–545 (2003).
Lang, R. et al. SOCS3 regulates the plasticity of gp130 signaling. Nat. Immunol. 4, 546–550 (2003).
Yang, X.O. et al. The signaling suppressor CIS controls proallergic T cell development and allergic airway inflammation. Nat. Immunol. 14, 732–740 (2013).
Takahashi, R. et al. SOCS1 is essential for regulatory T cell functions by preventing loss of Foxp3 expression as well as IFN- and IL-17A production. J. Exp. Med. 208, 2055–2067 (2011).
Luckey, M.A. et al. The transcription factor ThPOK suppresses Runx3 and imposes CD4+ lineage fate by inducing the SOCS suppressors of cytokine signaling. Nat. Immunol. 15, 638–645 (2014).
Spence, S. et al. Suppressors of cytokine signaling 2 and 3 diametrically control macrophage polarization. Immunity 38, 66–78 (2012).
Pellegrini, M. et al. IL-7 engages multiple mechanisms to overcome chronic viral infection and limit organ pathology. Cell 144, 601–613 (2011).
Delconte, R.B. et al. The helix-loop-helix protein ID2 governs NK cell fate by tuning their sensitivity to interleukin-15. Immunity 44, 103–115 (2016).
Miyagi, T. et al. High basal STAT4 balanced by STAT1 induction to control type 1 interferon effects in natural killer cells. J. Exp. Med. 204, 2383–2396 (2007).
Gil, M.P. et al. Regulating type 1 IFN effects in CD8 T cells during viral infections: changing STAT4 and STAT1 expression for function. Blood 120, 3718–3728 (2012).
Villarino, A.V. et al. Signal transducer and activator of transcription 5 (STAT5) paralog dose governs T cell effector and regulatory functions. eLife http://dx.doi.org/10.7554/eLife.08384 (2016).
Usui, T., Nishikomori, R., Kitani, A. & Strober, W. GATA-3 suppresses Th1 development by downregulation of Stat4 and not through effects on IL-12Rβ2 chain or T-bet. Immunity 18, 415–428 (2003).
Cheon, H., Yang, J. & Stark, G.R. The functions of signal transducers and activators of transcriptions 1 and 3 as cytokine-inducible proteins. J. Interferon Cytokine Res. 31, 33–40 (2011).
Metser, G. et al. An autoregulatory enhancer controls mammary-specific STAT5 functions. Nucleic Acids Res. 44, 1052–1063 (2016).
Witte, S. & Muljo, S.A. Integrating non-coding RNAs in JAK-STAT regulatory networks. JAK-STAT 3, e28055 (2014).
Kim, T.K. & Maniatis, T. Regulation of interferon-gamma-activated STAT1 by the ubiquitin-proteasome pathway. Science 273, 1717–1719 (1996).
Tanaka, T., Soriano, M.A. & Grusby, M.J. SLIM is a nuclear ubiquitin E3 ligase that negatively regulates STAT signaling. Immunity 22, 729–736 (2005).
Tanaka, T. et al. PDLIM2 inhibits T helper 17 cell development and granulomatous inflammation through degradation of STAT3. Sci. Signal. 4, ra85 (2011).
Sahoo, A., Alekseev, A., Obertas, L. & Nurieva, R. Grail controls Th2 cell development by targeting STAT6 for degradation. Nat. Commun. 5, 4732 (2014).
Villarino, A.V., Kanno, Y., Ferdinand, J.R. & O'Shea, J.J. Mechanisms of Jak/STAT signaling in immunity and disease. J. Immunol. 194, 21–27 (2015).
Delgoffe, G.M. & Vignali, D.A. STAT heterodimers in immunity. JAK-STAT 2, e23060 (2014).
Vinkemeier, U., Moarefi, I., Darnell, J.E. Jr. & Kuriyan, J. Structure of the amino-terminal protein interaction domain of STAT-4. Science 279, 1048–1052 (1998).
Begitt, A. et al. STAT1-cooperative DNA binding distinguishes type 1 from type 2 interferon signaling. Nat. Immunol. 15, 168–176 (2014).
Lin, J.-X. et al. Critical role of STAT5 transcription factor tetramerization for cytokine responses and normal immune function. Immunity 36, 586–599 (2012).
Yang, J. et al. Unphosphorylated STAT3 accumulates in response to IL-6 and activates transcription by binding to NFkappaB. Genes Dev. 21, 1396–1408 (2007).
Cheon, H. & Stark, G.R. Unphosphorylated STAT1 prolongs the expression of interferon-induced immune regulatory genes. Proc. Natl. Acad. Sci. USA 106, 9373–9378 (2009).
Park, H.J. et al. Cytokine-induced megakaryocytic differentiation is regulated by genome-wide loss of a uSTAT transcriptional program. EMBO J. 35, 580–594 (2015).
Carbognin, E., Betto, R.M., Soriano, M.E., Smith, A.G. & Martello, G. Stat3 promotes mitochondrial transcription and oxidative respiration during maintenance and induction of naive pluripotency. EMBO J. 35, 618–634 (2016).
Putz, E.M. et al. CDK8-mediated STAT1–S727 phosphorylation restrains NK cell cytotoxicity and tumor surveillance. Cell Rep. 4, 437–444 (2013).
Bancerek, J. et al. CDK8 kinase phosphorylates transcription factor STAT1 to selectively regulate the interferon response. Immunity 10.1016/j.immuni. 38, 250–262 (2013).
Begitt, A., Droescher, M., Knobeloch, K.P. & Vinkemeier, U. SUMO conjugation of STAT1 protects cells from hyperresponsiveness to IFN. Blood 118, 1002–1007 (2011).
Wegrzyn, J. et al. Function of mitochondrial Stat3 in cellular respiration. Science 323, 793–797 (2009).
Gough, D.J. et al. Mitochondrial STAT3 supports Ras-Dependent oncogenic transformation. Science 324, 1713–1716 (2009).
Friedbichler, K. et al. Stat5a serine 725 and 779 phosphorylation is a prerequisite for hematopoietic transformation. Blood 116, 1548–1558 (2010).
Gough, D.J., Marié, I.J., Lobry, C., Aifantis, I. & Levy, D.E. STAT3 supports experimental K-RasG12D-induced murine myeloproliferative neoplasms dependent on serine phosphorylation. Blood 124, 2252–2261 (2014).
Dasgupta, M., Dermawan, J.K.T., Willard, B. & Stark, G.R. STAT3-driven transcription depends upon the dimethylation of K49 by EZH2. Proc. Natl. Acad. Sci. USA 112, 3985–3990 (2015).
Beier, U.H. et al. Histone deacetylases 6 and 9 and sirtuin-1 control Foxp3+ regulatory T cell function through shared and isoform-specific mechanisms. Sci. Signal. 5, ra45 (2012).
Yarilina, A., Park-Min, K.-H., Antoniv, T., Hu, X. & Ivashkiv, L.B. TNF activates an IRF1-dependent autocrine loop leading to sustained expression of chemokines and STAT1-dependent type I interferon–response genes. Nat. Immunol. 9, 378–387 (2008).
Ivashkiv, L.B. & Donlin, L.T. Regulation of type I interferon responses. Nat. Rev. Immunol. 14, 36–49 (2014).
Bezbradica, J.S., Rosenstein, R.K., Demarco, R.A., Brodsky, I. & Medzhitov, R. A role for the ITAM signaling module in specifying cytokine-receptor functions. Nat. Immunol. 15, 333–342 (2014).
Yi, Z., Lin, W.W., Stunz, L.L. & Bishop, G.A. The adaptor TRAF3 restrains the lineage determination of thymic regulatory T cells by modulating signaling via the receptor for IL-2. Nat. Immunol. 15, 866–874 (2014).
Nagashima, H. et al. The adaptor TRAF5 limits the differentiation of inflammatory CD4+ T cells by antagonizing signaling via the receptor for IL-6. Nat. Immunol. 15, 449–456 (2014).
Upton, C., Mossman, K. & McFadden, G. Encoding of a homolog of the IFN-γ receptor by myxoma virus. Science 258, 1369–1372 (1992).
Guan, K.L., Broyles, S.S. & Dixon, J.E.A. Tyr/Ser protein phosphatase encoded by vaccinia virus. Nature 350, 359–362 (1991).
Morrison, T.E., Mauser, A., Wong, A., Ting, J.P. & Kenney, S.C. Inhibition of IFN-γ signaling by an Epstein-Barr virus immediate-early protein. Immunity 15, 787–799 (2001).
Ong, Y.C., Reese, M.L. & Boothroyd, J.C. Toxoplasma rhoptry protein 16 (ROP16) subverts host function by direct tyrosine phosphorylation of STAT6. J. Biol. Chem. 285, 28731–28740 (2010).
Saeij, J.P.J. et al. Toxoplasma co-opts host gene expression by injection of a polymorphic kinase homologue. Nature 445, 324–327 (2006).
Olias, P., Etheridge, R.D., Zhang, Y., Holtzman, M.J. & Sibley, L.D. Toxoplasma effector recruits the Mi-2/NuRD complex to repress STAT1 Transcription and block IFN-γ-dependent gene expression. Cell Host Microbe 20, 72–82 (2016).
Gay, G. et al. Toxoplasma gondii TgIST co-opts host chromatin repressors dampening STAT1-dependent gene regulation and IFN-γ-mediated host defenses. J. Exp. Med. 213, 1779–1798 (2016).
Casanova, J.L., Holland, S.M. & Notarangelo, L.D. Inborn errors of Human JAKs and STATs. Immunity 36, 515–528 (2012).
Kanai, T., Jenks, J. & Nadeau, K.C. The STAT5b pathway defect and autoimmunity. Front. Immunol. 3, 234 (2012).
Boisson-Dupuis, S. et al. Inborn errors of human STAT1: allelic heterogeneity governs the diversity of immunological and infectious phenotypes. Curr. Opin. Immunol. 24, 364–378 (2012).
Chen, E., Staudt, L.M. & Green, A.R. Janus kinase deregulation in leukemia and lymphoma. Immunity 36, 529–541 (2012).
Robinson, D.R. et al. Identification of recurrent NAB2-STAT6 gene fusions in solitary fibrous tumor by integrative sequencing. Nat. Genet. 45, 180–185 (2013).
Chmielecki, J. et al. Whole-exome sequencing identifies a recurrent NAB2-STAT6 fusion in solitary fibrous tumors. Nat. Genet. 45, 131–132 (2013).
Dong, S. & Tweardy, D.J. Interactions of STAT5b-RARα, a novel acute promyelocytic leukemia fusion protein, with retinoic acid receptor and STAT3 signaling pathways. Blood 99, 2637–2646 (2002).
Pilati, C. et al. Somatic mutations activating STAT3 in human inflammatory hepatocellular adenomas. J. Exp. Med. 208, 1359–1366 (2011).
Koskela, H.L.M. et al. Somatic STAT3 mutations in large granular lymphocytic leukemia. N. Engl. J. Med. 366, 1905–1913 (2012).
Jerez, A. et al. STAT3 mutations unify the pathogenesis of chronic lymphoproliferative disorders of NK cells and T-cell large granular lymphocyte leukemia. Blood 120, 3048–3057 (2012).
Rajala, H.L.M. et al. Discovery of somatic STAT5b mutations in large granular lymphocytic leukemia. Blood 121, 4541–4550 (2013).
Yildiz, M. et al. Activating STAT6 mutations in follicular lymphoma. Blood 125, 668–679 (2014).
Kiel, M.J. et al. Genomic analyses reveal recurrent mutations in epigenetic modifiers and the JAK-STAT pathway in Sezary syndrome. Nat. Commun. 6, 8470 (2015).
Crescenzo, R. et al. Convergent mutations and kinase fusions lead to oncogenic STAT3 activation in anaplastic large cell lymphoma. Cancer Cell 27, 516–532 (2015).
Bettelli, E. et al. Loss of T-bet, but not STAT1, prevents the development of experimental autoimmune encephalomyelitis. J. Exp. Med. 200, 79–87 (2004).
Villarino, A.V., Gallo, E. & Abbas, A.K. STAT1-activating cytokines limit Th17 responses through both T-bet-dependent and -independent mechanisms. J. Immunol. 185, 6461–6471 (2010).
Takeda, K. et al. Targeted disruption of the mouse Stat3 gene leads to early embryonic lethality. Proc. Natl. Acad. Sci. USA 94, 3801–3804 (1997).
Stritesky, G.L. et al. The transcription factor STAT3 is required for T helper 2 cell development. Immunity 34, 39–49 (2011).
Zhou, L. et al. IL-6 programs TH-17 cell differentiation by promoting sequential engagement of the IL-21 and IL-23 pathways. Nat. Immunol. 8, 967–974 (2007).
Ray, J.P. et al. Transcription factor STAT3 and type I interferons are corepressive insulators for differentiation of follicular helper and T helper 1 cells. Immunity 40, 367–377 (2014).
Cui, W., Liu, Y., Weinstein, J.S., Craft, J. & Kaech, S.M. An interleukin-21- interleukin-10-STAT3 pathway is critical for functional maturation of memory CD8+ T cells. Immunity 35, 792–805 (2011).
Guo, X. et al. Induction of innate lymphoid cell-derived interleukin-22 by the transcription factor STAT3 mediates protection against intestinal infection. Immunity 40, 25–39 (2014).
Laouar, Y., Welte, T., Fu, X.-Y. & Flavell, R.A. STAT3 is required for Flt3L-dependent dendritic cell differentiation. Immunity 19, 903–912 (2003).
Cheng, F. et al. A critical role for Stat3 signaling in immune tolerance. Immunity 19, 425–436 (2003).
Takeda, K. et al. Enhanced Th1 activity and development of chronic enterocolitis in mice devoid of Stat3 in macrophages and neutrophils. Immunity 10, 39–49 (1999).
Yao, Z. et al. Stat5a/b are essential for normal lymphoid development and differentiation. Proc. Natl. Acad. Sci. USA 103, 1000–1005 (2006).
Yao, Z. et al. Nonredundant roles for Stat5a/b in directly regulating Foxp3. Blood 109, 4368–4375 (2007).
Laurence, A. et al. Interleukin-2 signaling via STAT5 constrains T helper 17 cell generation. Immunity 26, 371–381 (2007).
Liao, W., Lin, J.-X., Wang, L., Li, P. & Leonard, W.J. Modulation of cytokine receptors by IL-2 broadly regulates differentiation into helper T cell lineages. Nat. Immunol. 12, 551–559 (2011).
Liao, W. et al. Priming for T helper type 2 differentiation by interleukin 2–mediated induction of interleukin 4 receptor α-chain expression. Nat. Immunol. 9, 1288–1296 (2008).
Eckelhart, E. et al. A novel Ncr1-Cre mouse reveals the essential role of STAT5 for NK-cell survival and development. Blood 117, 1565–1573 (2011).
Mitchell, D.M. & Williams, M.A. Disparate roles for STAT5 in primary and secondary CTL responses. J. Immunol. 190, 3390–3398 (2013).
Esashi, E. et al. The signal transducer STAT5 inhibits plasmacytoid dendritic cell development by suppressing transcription factor IRF8. Immunity 28, 509–520 (2008).
Bell, B.D. et al. The transcription factor STAT5 is critical in dendritic cells for the development of TH2 but not TH1 responses. Nat. Immunol. 14, 1–10 (2013).
Tiedt, R. et al. Ratio of mutant JAK2–V617F to wild-type Jak2 determines the MPD phenotypes in transgenic mice. Blood 111, 3931–3940 (2008).
Steward-Tharp, S.M. et al. A mouse model of HIES reveals pro- and anti-inflammatory functions of STAT3. Blood 123, 2978–2987 (2014).
Ohtani, T. et al. Dissection of signaling cascades through gp130 in vivo. Immunity 12, 95–105 (2000).
Jenkins, B.J. et al. Hyperactivation of Stat3 in gp130 mutant mice promotes gastric hyperproliferation and desensitizes TGF-β signaling. Nat. Med. 11, 845–852 (2005).
Varinou, L. et al. Phosphorylation of the Stat1 transactivation domain is required for full-fledged IFN-gamma-dependent innate immunity. Immunity 19, 793–802 (2003).
Dixit, A. et al. Perturb-Seq: dissecting molecular circuits with scalable single-cell RNA profiling of pooled genetic screens. Cell 167, 1853–1857 (2016).
Jaitin, D.A. et al. Dissecting immune circuits by linking CRISPR-pooled screens with single-cell RNA-seq. Cell 167, 1883–1896 (2016).
O'Shea, J.J., Kanno, Y. & Chan, A.C. In search of magic bullets: the golden age of immunotherapeutics. Cell 157, 227–240 (2014).
Various. Recombinant IL-2, IL-7, IL-12 and IL-15 clinical trials (US). ClinicalTrials.gov. https://clinicaltrials.gov/ct2/results?term=&recr=&type=Intr&rslt=&age_v=&gndr=&cond=&intr=Recombinant+AND+IL-2+OR+IL-7+OR+IL-15+OR+IL-12 (2016).
Arenas-Ramirez, N., Woytschak, J. & Boyman, O. Interleukin-2: biology, design and application. Trends Immunol. 36, 763–777 (2015).
Neri, D. & Sondel, P.M. ScienceDirectImmunocytokines for cancer treatment: past, present and future. Autoimmunity 40, 96–102 (2016).
Lefranc, M.-P. Immunoglobulin and T cell receptor genes: IMGT® and the birth and rise of immunoinformatics. Front. Immunol. 5, 22 (2014).
Toffalini, F. & Demoulin, J.-B. New insights into the mechanisms of hematopoietic cell transformation by activated receptor tyrosine kinases. Blood 116, 2429–2437 (2010).
Sen, M. & Grandis, J.R. Nucleic acid-based approaches to STAT inhibition. JAK-STAT 1, 285–291 (2012).
Miklossy, G., Hilliard, T.S. & Turkson, J. Therapeutic modulators of STAT signalling for human diseases. Nat. Rev. Drug Discov. 12, 611–629 (2013).
Various. STAT inhibitor clinical trials (US). ClinicalTrials.gov. Available at: https://clinicaltrials.gov/ct2/results?term=&recr=&type=Intr&rslt=&age_v=&gndr=&cond=&intr=STAT3+OR+STAT5; accessed 21 December (2016).
Acknowledgements
We thank members of the O'Shea laboratory for discussions about Jak–STAT biology, and C. Hunter and L. Hennighausen for critical reading of this manuscript. Supported by the National Institute of Arthritis, Musculoskeletal and Skin Diseases and the US National Institutes of Health (intramural funding).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
J.J.O. and the National Institutes of Health hold patents related to Jaks as targets of immunomodulatory agents and have a Collaborative Research Agreement and Development Award with Pfizer.
Supplementary information
Supplementary Text and Figures
Supplementary Table 1 (PDF 161 kb)
Rights and permissions
About this article
Cite this article
Villarino, A., Kanno, Y. & O'Shea, J. Mechanisms and consequences of Jak–STAT signaling in the immune system. Nat Immunol 18, 374–384 (2017). https://doi.org/10.1038/ni.3691
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ni.3691
This article is cited by
-
Activating mutations in JAK2 and CALR differentially affect intracellular calcium flux in store operated calcium entry
Cell Communication and Signaling (2024)
-
A Review on Nanosystem-Based Delivery of Tofacitinib for Enhanced Treatment of Autoimmune Diseases and Inflammation
BioNanoScience (2024)
-
Taming the cytokine storm: small molecule inhibitors targeting IL-6/IL-6α receptor
Molecular Diversity (2024)
-
Integrative function of histone deacetylase 3 in inflammation
Molecular Biology Reports (2024)
-
Does baricitinib reduce disease activity in patients with systemic lupus erythematosus? A systematic review and meta-analysis of randomized controlled trials
Clinical Rheumatology (2024)