Abstract
The cellular sources of interleukin 6 (IL-6) that are relevant for differentiation of the TH17 subset of helper T cells remain unclear. Here we used a novel strategy for the conditional deletion of distinct IL-6-producing cell types to show that dendritic cells (DCs) positive for the signaling regulator Sirpα were essential for the generation of pathogenic TH17 cells. Using their IL-6 receptor α-chain (IL-6Rα), Sirpα+ DCs trans-presented IL-6 to T cells during the process of cognate interaction. While ambient IL-6 was sufficient to suppress the induction of expression of the transcription factor Foxp3 in T cells, trans-presentation of IL-6 by DC-bound IL-6Rα (called 'IL-6 cluster signaling' here) was needed to prevent premature induction of interferon-γ (IFN-γ) expression in T cells and to generate pathogenic TH17 cells in vivo. Our findings should guide therapeutic approaches for the treatment of TH17-cell-mediated autoimmune diseases.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Accession codes
Change history
23 January 2017
In the version of this article initially published, the label for third bar from the left in Figure 4f ('Anti-IL-6α') was incorrect; the correct label is 'Anti-IL-6Rα'. Also, in the legend for Figure 4f, the description of the treatment conditions for middle four bars ('medium alone, or LPS and anti-IL-6 (MR16-1 or polyclonal antibody), anti-IL-6R (mAb#8)') was incorrect; the correct description is 'LPS alone (Medium), or LPS and anti-IL-6Rα (MR16-1), anti-IL-6 (polyclonal antibody or mAb#8)'. The error has been corrected for the print, PDF and HTML versions of this article.
References
Hall, A.O. et al. The cytokines interleukin 27 and interferon-γ promote distinct Treg cell populations required to limit infection-induced pathology. Immunity 37, 511–523 (2012).
Neufert, C. et al. IL-27 controls the development of inducible regulatory T cells and Th17 cells via differential effects on STAT1. Eur. J. Immunol. 37, 1809–1816 (2007).
Huber, M. et al. IL-27 inhibits the development of regulatory T cells via STAT3. Int. Immunol. 20, 223–234 (2008).
Dardalhon, V. et al. IL-4 inhibits TGF-β-induced Foxp3+ T cells and, together with TGF-β, generates IL-9+IL-10+Foxp3− effector T cells. Nat. Immunol. 9, 1347–1355 (2008).
Bettelli, E. et al. Reciprocal developmental pathways for the generation of pathogenic effector TH17 and regulatory T cells. Nature 441, 235–238 (2006).
Serada, S. et al. IL-6 blockade inhibits the induction of myelin antigen-specific Th17 cells and Th1 cells in experimental autoimmune encephalomyelitis. Proc. Natl. Acad. Sci. USA 105, 9041–9046 (2008).
Ghoreschi, K. et al. Generation of pathogenic TH17 cells in the absence of TGF-β signalling. Nature 467, 967–971 (2010).
Lee, Y. et al. Induction and molecular signature of pathogenic TH17 cells. Nat. Immunol. 13, 991–999 (2012).
Gaublomme, J.T. et al. Single-cell genomics unveils critical regulators of Th17 cell pathogenicity. Cell 163, 1400–1412 (2015).
Boulanger, M.J., Chow, D.-C., Brevnova, E.E. & Garcia, K.C. Hexameric structure and assembly of the interleukin-6/IL-6 alpha-receptor/gp130 complex. Science 300, 2101–2104 (2003).
Babon, J.J., Varghese, L.N. & Nicola, N.A. Inhibition of IL-6 family cytokines by SOCS3. Semin. Immunol. 26, 13–19 (2014).
Tamura, T. et al. Soluble interleukin-6 receptor triggers osteoclast formation by interleukin 6. Proc. Natl. Acad. Sci. USA 90, 11924–11928 (1993).
Rose-John, S. & Heinrich, P.C. Soluble receptors for cytokines and growth factors: generation and biological function. Biochem. J. 300, 281–290 (1994).
Jostock, T. et al. Soluble gp130 is the natural inhibitor of soluble interleukin-6 receptor transsignaling responses. Eur. J. Biochem. 268, 160–167 (2001).
Korn, T. et al. IL-21 initiates an alternative pathway to induce proinflammatory TH17 cells. Nature 448, 484–487 (2007).
Durant, L. et al. Diverse targets of the transcription factor STAT3 contribute to T cell pathogenicity and homeostasis. Immunity 32, 605–615 (2010).
Baran, P., Nitz, R., Grötzinger, J., Scheller, J. & Garbers, C. Minimal interleukin 6 (IL-6) receptor stalk composition for IL-6 receptor shedding and IL-6 classic signaling. J. Biol. Chem. 288, 14756–14768 (2013).
Palacios, R. & Steinmetz, M. Il-3-dependent mouse clones that express B-220 surface antigen, contain Ig genes in germ-line configuration, and generate B lymphocytes in vivo. Cell 41, 727–734 (1985).
Fischer, M. et al. I. A bioactive designer cytokine for human hematopoietic progenitor cell expansion. Nat. Biotechnol. 15, 142–145 (1997).
White, J. et al. The V beta-specific superantigen staphylococcal enterotoxin B: stimulation of mature T cells and clonal deletion in neonatal mice. Cell 56, 27–35 (1989).
Söderberg, O. et al. Direct observation of individual endogenous protein complexes in situ by proximity ligation. Nat. Methods 3, 995–1000 (2006).
Linker, R.A. et al. IL-6 transsignalling modulates the early effector phase of EAE and targets the blood-brain barrier. J. Neuroimmunol. 205, 64–72 (2008).
Nurieva, R. et al. Essential autocrine regulation by IL-21 in the generation of inflammatory T cells. Nature 448, 480–483 (2007).
Suto, A., Wurster, A.L., Reiner, S.L. & Grusby, M.J. IL-21 inhibits IFN-γ production in developing Th1 cells through the repression of Eomesodermin expression. J. Immunol. 177, 3721–3727 (2006).
Yagi, R. et al. The transcription factor GATA3 actively represses RUNX3 protein-regulated production of interferon-γ. Immunity 32, 507–517 (2010).
Awasthi, A. et al. Cutting edge: IL-23 receptor gfp reporter mice reveal distinct populations of IL-17-producing cells. J. Immunol. 182, 5904–5908 (2009).
Chu, C.Q., Wittmer, S. & Dalton, D.K. Failure to suppress the expansion of the activated CD4 T cell population in interferon γ-deficient mice leads to exacerbation of experimental autoimmune encephalomyelitis. J. Exp. Med. 192, 123–128 (2000).
Dubois, S., Mariner, J., Waldmann, T.A. & Tagaya, Y. IL-15Rα recycles and presents IL-15 In trans to neighboring cells. Immunity 17, 537–547 (2002).
Plun-Favreau, H. et al. The ciliary neurotrophic factor receptor α component induces the secretion of and is required for functional responses to cardiotrophin-like cytokine. EMBO J. 20, 1692–1703 (2001).
Wuest, S.C. et al. A role for interleukin-2 trans-presentation in dendritic cell-mediated T cell activation in humans, as revealed by daclizumab therapy. Nat. Med. 17, 604–609 (2011).
Tussiwand, R. & Gautier, E.L. Transcriptional regulation of mononuclear phagocyte development. Front. Immunol. 6, 533 (2015).
Lewis, K.L. et al. Notch2 receptor signaling controls functional differentiation of dendritic cells in the spleen and intestine. Immunity 35, 780–791 (2011).
Croxford, A.L., Mair, F. & Becher, B. IL-23: one cytokine in control of autoimmunity. Eur. J. Immunol. 42, 2263–2273 (2012).
Tanaka, T. et al. Enhancement of T helper2 response in the absence of interleukin (IL-)6; an inhibition of IL-4-mediated T helper2 cell differentiation by IL-6. Cytokine 13, 193–201 (2001).
Diehl, S. & Rincón, M. The two faces of IL-6 on Th1/Th2 differentiation. Mol. Immunol. 39, 531–536 (2002).
Kim, J.H. et al. High cleavage efficiency of a 2A peptide derived from porcine teschovirus-1 in human cell lines, zebrafish and mice. PLoS One 6, e18556 (2011).
Quintana, A. et al. Astrocyte-specific deficiency of interleukin-6 and its receptor reveal specific roles in survival, body weight and behavior. Brain Behav. Immun. 27, 162–173 (2013).
Korn, T. et al. Myelin-specific regulatory T cells accumulate in the CNS but fail to control autoimmune inflammation. Nat. Med. 13, 423–431 (2007).
Bettelli, E. et al. Myelin oligodendrocyte glycoprotein-specific T cell receptor transgenic mice develop spontaneous autoimmune optic neuritis. J. Exp. Med. 197, 1073–1081 (2003).
Wunderlich, F.T. et al. Interleukin-6 signaling in liver-parenchymal cells suppresses hepatic inflammation and improves systemic insulin action. Cell Metab. 12, 237–249 (2010).
Rabe, B. et al. Transgenic blockade of interleukin 6 transsignaling abrogates inflammation. Blood 111, 1021–1028 (2008).
Korn, T. et al. IL-6 controls Th17 immunity in vivo by inhibiting the conversion of conventional T cells into Foxp3+ regulatory T cells. Proc. Natl. Acad. Sci. USA 105, 18460–18465 (2008).
Brakenhoff, J.P., Hart, M., De Groot, E.R., Di Padova, F. & Aarden, L.A. Structure-function analysis of human IL-6. Epitope mapping of neutralizing monoclonal antibodies with amino- and carboxyl-terminal deletion mutants. J. Immunol. 145, 561–568 (1990).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Anders, S., Pyl, P.T. & Huber, W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169 (2015).
Love, M.I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Schindelin, J. et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012).
Acknowledgements
We thank all members of the Korn laboratory for input; B. Kunze, B. Lunk and colleagues for mouse care; D. Busch (Technical University of Munich) for OT-II mice (Tg(TcraTcrb)425Cbn) expressing the congenic marker CD45.1; F. Greten (Georg-Speyer Haus) for Stat3flox/flox (Stat3tm2Aki) mice; B. Stockinger (MRC National Institute for Medical Research) for the 19E12 hybridoma; and M. Kopf (ETH, Zürich) for Il21r−/− mice. Supported by the Deutsche Forschungsgemeinschaft (TRR128 to A.W. and T.K.; TRR156 and WA1600/8-1 to A.W.; SFB1054-B07 and SyNergy to T.K.; SFB877-A01 to S.R.-J.; SFB877-A10 to C. Gar.; and the cluster of excellence 'Inflammation at interfaces' to S.R.-J. and C. Gar.), the European Research Council (Consolidator Grant 647215 to T.K.), and the Spanish Ministerio de Economía y Competitividad (SAF2011-23272 and SAF2014-56546-R to J.H.).
Author information
Authors and Affiliations
Contributions
S.H. designed, performed and analyzed most experiments and drafted the manuscript; N.Y. performed and analyzed key in vivo experiments; C. Gar., M.H., L.A., V.H., A.L.C., K.M.-H. and T.R. performed or contributed to specific experiments; C.Gas. performed bioinformatics analysis; H.S.B. and K.S. performed and analyzed nanostring experiments; S.K. and H.B. performed and analyzed RNA-sequencing experiments; B.H., T.M., T.F.W., J.H., M.O., S.R.-J., M.S.-S. provided reagents, advice, design and supervision of experiments; A.W. supervised experiments, analyzed data and wrote the manuscript; and T.K. conceptualized the study, designed and supervised the experiments, analyzed data and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
Supplementary Figure 1 IL-6 reporter mice.
(a) Targeted Il6 locus. The reporter cassette including a floxed stop cassette was introduced into exon 2 of the Il6 locus. Since the locus is disrupted by this knock-in construct, IL-6 reporter mice were bred heterozygously and compared with Il6+/- mice since Il6 produces a gene dose effect (data not shown). (b) Bone marrow derived dendritic cells (BMDCs) were prepared from CMV-Cre x Il6RD/wt mice and stimulated in vitro with CpG. Cerulean was expressed in the cytoplasm (left) and Thy1.1 at the cell surface (middle, merge right) as expected. Confocal microphotographs, Scale bar 10 μm. (c) Il6RD/wt control BMDCs or BMDCs prepared from IL-6 reporter mice (CMV- Cre x Il6RD/wt) were stimulated with LPS followed by flow cytometric assessment of Thy1.1 expression. (d-f) Correlation of Thy1.1 expression and IL-6 expression in BMDCs. BMDCs were prepared from different mouse strains as indicated and stimulated with LPS followed by analysis of Thy1.1 expression (d) and IL-6 production as measured by ELISA (e). (f) Co-expression of IL-6 and Thy1.1. Control BMDCs (Il6RD/wt) and IL-6 reporter BMDCs (CD11c-Cre x Il6RD/wt) were stimulated with CpG for 6 h in the presence of Brefeldin A for the last 2 h followed by combined surface staining for Thy1.1 and intracellular staining for IL-6.
Supplementary Figure 2 Characterization in vivo of DCs expressing IL-6 (Thy1.1).
(a) Il6RD/wt mice were crossed to CD11c-Cre and R26-Stopflox/flox-YFP mice to generate compound heterozygous mice with DC conditional expression of an IL-6 reporter allele and YFP. In order to visualize large amounts of IL-6 producing DCs ex vivo, we injected Flt3L producing melanoma cells s.c. to expand DCs in vivo and 6 days later, treated the animals with LPS (3 mg/kg LPS [E. coli 0111:B4]) i.p. to stimulate IL-6 production. Two days after LPS injection, lymph nodes (LN) and spleen (SPL) were prepared and stained for Thy1.1 to visualize IL-6+ DCs by flow cytometry directly ex vivo. In order to analyze whether Thy1.1 (IL-6)+ DCs segregate into a specific DC subset, the indicated surface molecules were co-stained. (b) IL-6 Reporter mice allow for IL-6 conditional deletion of DCs in vivo. CD11c-Cre x Il6RD/wt x R26-Stopflox/flox-YFP mice were treated with Flt3L producing melanoma cells and LPS. The mice were then assigned to treatment with either Isotype (mouse IgG2a, C1.18.4) or anti-Thy1.1 (19E12) antibody treatment in order to deplete IL-6+ DCs. One day later, lymph nodes (LN) and spleen (SPL) were prepared and stained for Thy1.1 (OX-7) to visualize IL-6+ DCs by flow cytometry directly ex vivo. CD11c-Cre x R26-Stopflox/flox-YFP x Il6wt/wt mice, which were treated identically as the DC conditional IL-6 reporter mice, are shown as a "negative" staining control for Thy1.1 (left).
Supplementary Figure 3 In vivo priming of T cell responses in the absence of IL-6-producing DCs.
DC conditional IL-6 reporter mice (CD11c-Cre x Il6RD/wt) were immunized with MOG(35-55) in CFA followed by control treatment (mouse IgG2a isotype) or anti-Thy1.1 (19E12) to deplete IL-6 (Thy1.1)+ DCs. Antibody treatment was performed by i.p. injection of 200 μg antibody every other day starting on day 1 after immunization. On day 7 after immunization, draining lymph nodes (LN) and spleen (SPL) were prepared and stained for Foxp3 to quantify the fraction of Tregs in the CD4+ T cell compartment (a). (b) Subsequent to PMA/ionomycin restimulation, LN CD4+ T cells were stained intracellularly for IL-17, GM-CSF, IFN-γ, and IL-10. (c, d) Antigen specific T cell responses were assessed by intracellular staining of CD40L (CD154) and cytokines in splenic CD4+ T cells of control-treated or IL-6+ DC-depleted mice after recall with MOG(35-55). Mean + SD (n=5 mice per genotype).
Supplementary Figure 4 Ablation of Il6 in B cells, T cells or macrophages does not result in resistance to MOG(35–55)-induced EAE.
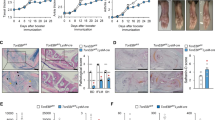
Course of MOG(35-55) induced EAE in mouse strains with conditional ablation of Il6 in B cells (CD19-Cre x Il6flox/flox, Il6ΔB) (a), T cells (CD4-Cre x Il6flox/flox, Il6ΔT) (b), and LysM+ myeloid cells (LysM-Cre x Il6flox/flox, Il6ΔM Φ) (c). Mice were subcutaneously immunized with MOG(35-55) in CFA and i.v. injected with pertussis toxin on days 0 and 2. Mean clinical EAE score and SEM, n ≥ 4 per group. *P<0.05, ANOVA plus Fisher's LSD test for individual days.
Supplementary Figure 5 Il6ra−/− BMDCs are not deficient in the production of pro-inflammatory cytokines in response to stimulation with LPS.
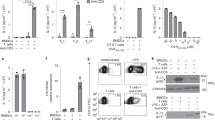
Control Il6raflox/flox or IL-6Rα deficient BMDCs (Il6raΔDC) were stimulated over night with either IL-6 or LPS followed by analysis of Il1b, Il6, Il12a, Il12b, and Il23 mRNA production by quantitative RT PCR. Mean + SD of technical replicates. One out of two independent experiments. *P<0.05, ANOVA plus Sidak’s multiple comparisons test.
Supplementary Figure 6 DC-derived IL-6 is required for robust activation of STAT3 in T cells.
(a, b) Subcutaneous immunization with a peptide antigen in CFA induces similar amounts of serum IL-6 in control mice (Il6flox/flox) and Il6ΔDC mice. Control animals (Il6flox/flox), Il6-/- mice, and Il6ΔDC mice were either injected with LPS (a) to induce systemic IL-6 or immunized subcutaneously with MOG(35-55) in CFA (b). Serum samples were collected 5 h after LPS injection or 1 day after subcutaneous immunization for the assessment of IL-6 by ELISA (n=3, SD, *P<0.04, One-way-ANOVA plus Tukey's multiple comparisons test). (c, d) RNA Seq analysis was performed in 2D2 T cells re-isolated from draining lymph nodes of control hosts (Il6flox/flox) or Il6ΔDC hosts after immunization with cognate MOG peptide. (c) Ingenuity pathway analysis was performed to evaluate the strength of STAT3 pathway activation in control primed (left panel) or Il6ΔDC primed (right panel) 2D2 effector T cells. (d) Notably, in contrast to T cells primed in a control milieu, Il6ΔDC primed T cells exhibited a weakened “STAT3” signature when their RNA profile was directly tested for the enrichment of STAT3 dependent genes by GSEA (see also Supplementary Tables).
Supplementary Figure 7 IL-6 cluster signaling and surface and intracellular expression of IL-6Rα and gp130 by CD11b+ DCs.
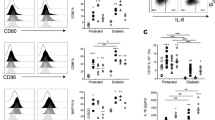
(a) Scheme of IL-6 cluster signaling. IL-6 is loaded onto the IL-6Rα in intracellular compartments of DCs and is brought to the cell membrane as an IL-6-IL-6Rα complex. During a cognate interaction between DCs and T cells, DCs present IL-6 via their IL-6Rα in trans to T cells. Trans-presentation of IL-6 leads to the engagement of gp130 on the T cell side (IL-6 cluster signaling) and induces a pathogenic phenotype in sensitized T cells. (b) CD11b+ DCs express IL-6Rα on their cell surface. WT mice were immunized with MOG(35-55) in CFA and on day 7 after immunization, cells from draining lymph nodes (LN) and spleen (SPL) were analyzed by flow cytometry. (b) IL-6Rα and gp130 expression was assessed in CD11c+MHC class II+CD11b+ cDC2 either by surface staining (left column) or by intracellular staining (right column). Grey: isotype. Blue overlay: IL-6Rα or gp130, respectively. (c) Surface expression of IL-6Rα was assessed on splenic cDC1 cells (CD103+) or CD11b+ DCs isolated from immunized mice on day 7 after immunization.
Supplementary Figure 8 IL-6 trans-signaling by the soluble IL-6–IL-6Rα complex is irrelevant during MOG(35–55)-induced EAE.
WT mice, opt_sgp130-Fc transgenic mice, and Il6-/- mice were immunized with MOG(35-55) in CFA. Opt_sgp130-Fc transgenic mice produce large amounts of sgp130, which blocks endogenous IL-6 trans-signaling in vivo. Mean EAE scores + SEM, *P<0.04, ANOVA plus Tukey post test.
Supplementary Figure 9 IL-6Rα-deficient T cells differentiate into pathogenic TH17 cells.
Il6raflox/flox control mice and Treg sufficient or deficient (anti-CD25 treated) Il6raΔT animals were immunized with MOG(35-55) in CFA. After priming of antigen specific T cells in vivo, the cytokine response was assessed in CD4+ T cells isolated from the spleen on day 10 after immunization after short term ex vivo restimulation with PMA/ionomycin and intracellular cytokine staining. (a) Representative cytograms of the CD4+ T cell gate. (b) Frequency of IFN-γ producing CD4+ T cells (left) and IL-17 producing CD4+ T cells (right); n=3 (Ctrl), n=4 (isotype treated Il6raΔT) and n=4 (anti-CD25 treated Il6raΔT). ANOVA, Fisher's LSD post-test, *P<0.03.
Supplementary Figure 10 IL-6 cluster signaling is sufficient to induce pathogenic TH17 cells in the simultaneous absence of classic IL-6 signaling and the IL-21-mediated alternative pathway for the induction of TH17 cells.
Naive CD4+ T cells were purified from Il6raflox/flox control mice, Il6raΔT mice, Il21r-/- mice, or Il6raΔT x Il21r-/- mice and transferred into Rag1-/- host animals followed by immunization with MOG(35-55) in CFA. (a) Intracellular cytokine staining of T cells re-isolated from the spleen on day 14 after immunization and subjected to ex vivo stimulation with PMA/ionomycin. (b) Clinical course of EAE in Rag1-/- recipients of T cells deficient in both IL-6Rα and IL-21R. Mean clinical score + SEM, n=3.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–10 and Supplementary Tables 1–4 (PDF 3009 kb)
Rights and permissions
About this article
Cite this article
Heink, S., Yogev, N., Garbers, C. et al. Trans-presentation of IL-6 by dendritic cells is required for the priming of pathogenic TH17 cells. Nat Immunol 18, 74–85 (2017). https://doi.org/10.1038/ni.3632
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ni.3632
This article is cited by
-
Targeting IL-6 or IL-6 Receptor in Rheumatoid Arthritis: What Have We Learned?
BioDrugs (2024)
-
Causal effect of interleukin (IL)-6 on blood pressure and hypertension: A mendelian randomization study
Immunogenetics (2024)
-
The Role of IL-6 in Neurodegenerative Disorders
Neurochemical Research (2024)
-
A discrete ‘early-responder’ stromal-cell subtype orchestrates immunocyte recruitment to injured tissue
Nature Immunology (2023)
-
Blockade of IL-6 signaling alleviates atherosclerosis in Tet2-deficient clonal hematopoiesis
Nature Cardiovascular Research (2023)