Abstract
TET proteins oxidize 5-methylcytosine in DNA to 5-hydroxymethylcytosine and other oxidation products. We found that simultaneous deletion of Tet2 and Tet3 in mouse CD4+CD8+ double-positive thymocytes resulted in dysregulated development and proliferation of invariant natural killer T cells (iNKT cells). Tet2-Tet3 double-knockout (DKO) iNKT cells displayed pronounced skewing toward the NKT17 lineage, with increased DNA methylation and impaired expression of genes encoding the key lineage-specifying factors T-bet and ThPOK. Transfer of purified Tet2-Tet3 DKO iNKT cells into immunocompetent recipient mice resulted in an uncontrolled expansion that was dependent on the nonclassical major histocompatibility complex (MHC) protein CD1d, which presents lipid antigens to iNKT cells. Our data indicate that TET proteins regulate iNKT cell fate by ensuring their proper development and maturation and by suppressing aberrant proliferation mediated by the T cell antigen receptor (TCR).
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Accession codes
References
Yui, M.A. & Rothenberg, E.V. Developmental gene networks: a triathlon on the course to T cell identity. Nat. Rev. Immunol. 14, 529–545 (2014).
Carpenter, A.C. & Bosselut, R. Decision checkpoints in the thymus. Nat. Immunol. 11, 666–673 (2010).
Bendelac, A., Savage, P.B. & Teyton, L. The biology of NKT cells. Annu. Rev. Immunol. 25, 297–336 (2007).
Vahedi, G. et al. Helper T-cell identity and evolution of differential transcriptomes and epigenomes. Immunol. Rev. 252, 24–40 (2013).
Hogquist, K.A. & Jameson, S.C. The self-obsession of T cells: how TCR signaling thresholds affect fate 'decisions' and effector function. Nat. Immunol. 15, 815–823 (2014).
Lee, Y.J., Holzapfel, K.L., Zhu, J., Jameson, S.C. & Hogquist, K.A. Steady-state production of IL-4 modulates immunity in mouse strains and is determined by lineage diversity of iNKT cells. Nat. Immunol. 14, 1146–1154 (2013).
Winter, D.R. & Amit, I. The role of chromatin dynamics in immune cell development. Immunol. Rev. 261, 9–22 (2014).
Smith, Z.D. & Meissner, A. DNA methylation: roles in mammalian development. Nat. Rev. Genet. 14, 204–220 (2013).
Tahiliani, M. et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science 324, 930–935 (2009).
Ito, S. et al. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science 333, 1300–1303 (2011).
He, Y.F. et al. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science 333, 1303–1307 (2011).
Pastor, W.A., Aravind, L. & Rao, A. TETonic shift: biological roles of TET proteins in DNA demethylation and transcription. Nat. Rev. Mol. Cell Biol. 14, 341–356 (2013).
Tsagaratou, A. et al. Dissecting the dynamic changes of 5-hydroxymethylcytosine in T-cell development and differentiation. Proc. Natl. Acad. Sci. USA 111, E3306–E3315 (2014).
Ko, M. et al. Impaired hydroxylation of 5-methylcytosine in myeloid cancers with mutant TET2. Nature 468, 839–843 (2010).
Ko, M. et al. Ten-Eleven-Translocation 2 (TET2) negatively regulates homeostasis and differentiation of hematopoietic stem cells in mice. Proc. Natl. Acad. Sci. USA 108, 14566–14571 (2011).
Gu, T.P. et al. The role of Tet3 DNA dioxygenase in epigenetic reprogramming by oocytes. Nature 477, 606–610 (2011).
Lee, Y.J., Jameson, S.C. & Hogquist, K.A. Alternative memory in the CD8 T cell lineage. Trends Immunol. 32, 50–56 (2011).
Engel, I. et al. Innate-like functions of natural killer T cell subsets result from highly divergent gene programs. Nat. Immunol. 17, 728–739 (2016).
Yu, S. et al. The TCF-1 and LEF-1 transcription factors have cooperative and opposing roles in T cell development and malignancy. Immunity 37, 813–826 (2012).
Karo, J.M., Schatz, D.G. & Sun, J.C. The RAG recombinase dictates functional heterogeneity and cellular fitness in natural killer cells. Cell 159, 94–107 (2014).
Matthews, J.M., Lester, K., Joseph, S. & Curtis, D.J. LIM-domain-only proteins in cancer. Nat. Rev. Cancer 13, 111–122 (2013).
Ramsay, R.G. & Gonda, T.J. MYB function in normal and cancer cells. Nat. Rev. Cancer 8, 523–534 (2008).
Uddin, M.N. et al. Transcription factor Bcl11b sustains iNKT1 and iNKT2 cell programs, restricts iNKT17 cell program, and governs iNKT cell survival. Proc. Natl. Acad. Sci. USA 113, 7608–7613 (2016).
Mathew, R. et al. BTB-ZF factors recruit the E3 ligase cullin 3 to regulate lymphoid effector programs. Nature 491, 618–621 (2012).
Dose, M. et al. Intrathymic proliferation wave essential for Valpha14+ natural killer T cell development depends on c-Myc. Proc. Natl. Acad. Sci. USA 106, 8641–8646 (2009).
Carr, T. et al. The transcription factor lymphoid enhancer factor 1 controls invariant natural killer T cell expansion and TH2-type effector differentiation. J. Exp. Med. 212, 793–807 (2015).
Yue, X. et al. Control of Foxp3 stability through modulation of TET activity. J. Exp. Med. 213, 377–397 (2016).
Wolf, E., Lin, C.Y., Eilers, M. & Levens, D.L. Taming of the beast: shaping Myc-dependent amplification. Trends Cell Biol. 25, 241–248 (2014).
Lee, Y.J. et al. Lineage-specific effector signatures of invariant NKT cells are shared amongst γδ T, innate lymphoid, and TH cells. J. Immunol. 197, 1460–1470 (2016).
Fonseca-Pereira, D. et al. The neurotrophic factor receptor RET drives haematopoietic stem cell survival and function. Nature 514, 98–101 (2014).
Huang, Y., Pastor, W.A., Zepeda-Martínez, J.A. & Rao, A. The anti-CMS technique for genome-wide mapping of 5-hydroxymethylcytosine. Nat. Protoc. 7, 1897–1908 (2012).
Pastor, W.A. et al. Genome-wide mapping of 5-hydroxymethylcytosine in embryonic stem cells. Nature 473, 394–397 (2011).
Laurent, L. et al. Dynamic changes in the human methylome during differentiation. Genome Res. 20, 320–331 (2010).
Shen, Y. et al. A map of the cis-regulatory sequences in the mouse genome. Nature 488, 116–120 (2012).
An, J. et al. Acute loss of TET function results in aggressive myeloid cancer in mice. Nat. Commun. 6, 10071 (2015).
Kieffer-Kwon, K.R. et al. Interactome maps of mouse gene regulatory domains reveal basic principles of transcriptional regulation. Cell 155, 1507–1520 (2013).
Lara-Astiaso, D. et al. Immunogenetics. chromatin state dynamics during blood formation. Science 345, 943–949 (2014).
Buenrostro, J.D., Giresi, P.G., Zaba, L.C., Chang, H.Y. & Greenleaf, W.J. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods 10, 1213–1218 (2013).
Jeong, M. et al. Large conserved domains of low DNA methylation maintained by Dnmt3a. Nat. Genet. 46, 17–23 (2014).
Townsend, M.J. et al. T-bet regulates the terminal maturation and homeostasis of NK and Valpha14i NKT cells. Immunity 20, 477–494 (2004).
Engel, I., Zhao, M., Kappes, D., Taniuchi, I. & Kronenberg, M. The transcription factor Th-POK negatively regulates TH17 differentiation in Vα14i NKT cells. Blood 120, 4524–4532 (2012).
Stritesky, G.L., Jameson, S.C. & Hogquist, K.A. Selection of self-reactive T cells in the thymus. Annu. Rev. Immunol. 30, 95–114 (2012).
Dobenecker, M.W. et al. Coupling of T cell receptor specificity to natural killer T cell development by bivalent histone H3 methylation. J. Exp. Med. 212, 297–306 (2015).
Pereira, R.M. et al. Jarid2 is induced by TCR signaling and controls iNKT cell maturation. Nat. Commun. 5, 4540 (2014).
Rasmussen, K.D. & Helin, K. Role of TET enzymes in DNA methylation, development, and cancer. Genes Dev. 30, 733–750 (2016).
Ichiyama, K. et al. The methylcytosine dioxygenase Tet2 promotes DNA demethylation and activation of cytokine gene expression in T cells. Immunity 42, 613–626 (2015).
Sasidharan Nair, V., Song, M.H. & Oh, K.I. Vitamin C facilitates demethylation of the Foxp3 enhancer in a Tet-dependent manner. J. Immunol. 196, 2119–2131 (2016).
Huang, Y. et al. Distinct roles of the methylcytosine oxidases Tet1 and Tet2 in mouse embryonic stem cells. Proc. Natl. Acad. Sci. USA 111, 1361–1366 (2014).
Ko, M. et al. TET proteins and 5-methylcytosine oxidation in hematological cancers. Immunol. Rev. 263, 6–21 (2015).
Lee, P.P. et al. A critical role for Dnmt1 and DNA methylation in T cell development, function, and survival. Immunity 15, 763–774 (2001).
Heinen, A.P. et al. Improved method to retain cytosolic reporter protein fluorescence while staining for nuclear proteins. Cytometry A 85, 621–627 (2014).
Djuretic, I.M. et al. Transcription factors T-bet and Runx3 cooperate to activate Ifng and silence Il4 in T helper type 1 cells. Nat. Immunol. 8, 145–153 (2007).
Picelli, S. et al. Full-length RNA-seq from single cells using Smart-seq2. Nat. Protoc. 9, 171–181 (2014).
Huang, Y. et al. The behaviour of 5-hydroxymethylcytosine in bisulfite sequencing. PLoS One 5, e8888 (2010).
Tsagaratou, A. & Rao, A. TET proteins and 5-methylcytosine oxidation in the immune system. Cold Spring Harb. Symp. Quant. Biol. 78, 1–10 (2013).
Acknowledgements
We thank C. Kim, K. van Gunst, L. Nosworthy, D. Hinz and R. Simmons at the LJI Flow Cytometry Core for help with fluorescence-activated cell sorting; G. Seumois and J. Day at the LJI Functional Genomics Center for assistance with next-generation sequencing (Illumina HiSeq 2500); M. Kronenberg, I. Engel and C.-W. Lio (LJI) for discussions, the LJI Bioinformatics Core for routine analysis; Z. Mikulski and B. Kiosses at the LJI microscopy core, M. Chadwell at the LJI Histology core, and the Histology Core at the University of California at San Diego Moores Cancer Center; and R. Bosselut (National Cancer Institute) for pMX-ThPOK-IRES-GFP. Supported by US National Institutes of Health (R01 AI44432, CA151535 and R35CA210043), the Leukemia and Lymphoma Society (Translation Research Project grant 6187-12 to A.R.), the Academy of Finland Centre of Excellence in Molecular Systems Immunology and Physiology Research (H.L. and. S.R.), an Albert Billings Ruddock Professorship at Caltech (E.V.R.), the Cancer Research Institute (Irvington Institute postdoctoral fellowship to A.T.), the Fraternal Order of Eagles Fellow of the Damon Runyon Cancer Research Foundation (DRG-2069-11 to J.P.S.-B.) and the National Science Foundation (W.A.P.).
Author information
Authors and Affiliations
Contributions
A.R. and A.T. designed the study; A.T. performed all of the experiments, and A.R. and A.T. wrote the manuscript. E.G.-A. analyzed WGBS and ATAC-seq data sets under the supervision of L.C. and J.P.S.-B., respectively. S.R. analyzed RNA-seq and CMS-IP data sets under the supervision of H.L. S.T. helped with in vivo adoptive transfer experiments. W.A.P. generated the Tet3fl/fl mice. E.V.R. provided critical input and suggestions during the course of this study and helped write the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
Supplementary Figure 1 Characterization of Tet2-3 DKO mice
(a) Tet1, Tet2 and Tet3 mRNA expression assessed by quantitative RT-PCR. PolyA+ RNA was isolated from DP, CD4 SP and CD8 SP T cells from wild type (WT) and Tet2-/-Tet3fl/fl CD4cre (Tet2-3 DKO) mice. Relative mRNA expression levels were normalized to Gapdh.
(b) 5hmC levels in genomic DNA of thymocyte subsets from WT (n=2) and Tet2-3 DKO (n=2) mice, estimated by anti-CMS dot blot after bisulfite treatment of genomic DNA to convert 5hmC to CMS. One representative experiment out of 2 is shown.
(c) Haematoxylin and eosin staining of spleen, lung and liver sections from 6 week-old control (upper panel) and Tet2-3 DKO (lower panel) mice.
(d) iNKT cells in lymph nodes (LN) of WT versus Tet2-/-, Tet3 KO and Tet2-3 DKO iNKT cells as defined by  GalCer-CD1d and TCRβ staining.
GalCer-CD1d and TCRβ staining.
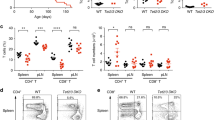
(e) Percentage (left) and number (right) of iNKT cells in lymph nodes. n=6 (WT), n=3 (Tet2 KO), n=at least 2 (Tet3 KO), n=6 (Tet2/3 DKO).
Data are mean ± SEM. *P<0.05, **P< 0.01, ***P< 0.001, ****P< 0.0001 (unpaired t test)
Supplementary Figure 2 Analysis of CD4 and CD8 T cell development in wild type and Tet2-3 DKO mice.
(a) Thymic cellularity in wild type (n=9) and Tet2-3 DKO (n=10) mice at 3-4 weeks of age.
(b) Top, Analysis of DN, DP, CD4 and CD8 SP thymic subsets defined by surface staining for CD4 and CD8 markers. Bottom, Representation of these subsets after gating out iNKT cells as defined by  GalCer-CD1d and TCRβ staining.
GalCer-CD1d and TCRβ staining.
(c) Percentage and (d), Number of DP, CD4 SP and CD8 SP cells in the thymus of WT (n=6) and Tet2-3 DKO (n=5) mice at 3-4 weeks.
(e) Representation of CD4 and CD8 T cells in lymph nodes (LN, top) and spleen (bottom) of WT and Tet2-3 DKO mice at 3-4 weeks.
(f) Total number of cells in lymph nodes (LN) of 3-4 week old WT (n=3) and Tet2-3 DKO (n=3) mice.
(g) Percentage and number of CD4 and CD8 cells in the LN of WT (n=6) and Tet2-3 DKO (n=3) mice analyzed at 3-4 weeks old.
(h) Percentage and number of CD4 and CD8 cells in the spleen of WT (n=6) and Tet2-3 DKO (n=3) mice analyzed at 3-4 weeks old.
Data are mean ± SEM. *P<0.05, **P< 0.01, ***P< 0.001, ****P< 0.0001 (unpaired t test).
At least 3 independent experiments were performed.
Supplementary Figure 3 Profound loss of TET proteins is required for dysregulated expansion and function of iNKT cells.
(a) iNKT cells in the thymus of representative 20 day-old WT versus Tet2-/-, Tet3 KO and Tet2-3 DKO mice, defined by staining with  GalCer-CD1d tetramer and anti-TCRβ.
GalCer-CD1d tetramer and anti-TCRβ.
(b) Increased percentages (left) and numbers (right) of iNKT cells in spleens isolated from 4 week-old WT (n=6), T2-/- (T2 KO, n=3), T3 KO (n=3) and Tet2-3 DKO (n=10) mice.
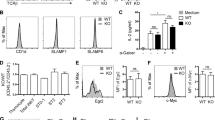
(c) Histogram evaluating the expression of RORγt (left); PLZF (center); and CD4 (right) in WT, single Tet2-/- (Tet2 KO), Tet3 KO, Tet2-3 DKO thymic iNKT cells.
(d, e) Histogram evaluating the expression of RORγt in the spleen (d) and lymph nodes (e) of WT, single Tet2-/-, Tet3 KO, Tet2-3 DKO iNKT cells.
Data are mean ± SEM. *P<0.05, **P< 0.01, ***P< 0.001, ****P< 0.0001 (unpaired t test)
Supplementary Figure 4 Cytokine production in Tet2-3 DKO iNKT cells.
(a) Representative flow cytometric analysis of IL17 secretion by  GalCer-CD1d tetramer+ TCRβ+ cells.
GalCer-CD1d tetramer+ TCRβ+ cells.
(b) Percentage of IL17-secreting iNKT cells among total iNKT cells.
(c) Percentage of IL17-secreting iNKT cells among total thymocytes.
(d) Representative flow cytometric analysis of IFNγ secretion by  GalCer-CD1d tetramer+ TCRβ+ TCR cells.
GalCer-CD1d tetramer+ TCRβ+ TCR cells.
(e) Percentage of IFNγ-secreting iNKT cells among total iNKT cells.
(f) Percentage of IFNγ-secreting iNKT cells among total thymocytes.
(g) Representative flow cytometric analysis of IL4 secretion by  GalCer-CD1d tetramer+ TCRβ+ cells.
GalCer-CD1d tetramer+ TCRβ+ cells.
(h) Percentage of IL4-secreting iNKT cells among iNKT cells.
(i) Percentage of IL4-secreting iNKT cells among total thymocytes.
In all cases (a-i) 3 mice per genotype were evaluated in 2 independent experiments.
(j) CD8 SP thymocytes were assessed by flow cytometry for the expression of markers that characterize memory-like CD8 cells: surface markers CD122, CD44, CXCR3 and the transcription factor Eomes.
(k) Percentage of CD8 SP thymocytes that express CD122, CD44, CXCR3 and Eomes. WT (n=3) and Tet2-3 DKO (n=3) mice. For (j) and (k) 3 independent experiments were performed.
Data are mean ± SEM. *P<0.05, **P< 0.01, ***P< 0.001, ****P< 0.0001 (unpaired t test)
Supplementary Figure 5 Increase of NKT17 subset and reduction of NKT1 precursors in the NK1.1- Tet2-3 DKO iNKT cells.
(a) Representative flow cytometric analysis of transcription factors PLZF and RORγt in  GalCer-CD1d tetramer+ CD24-TCRβ+ NK1.1- iNKT cells reveals dramatic increase of NKT17 subset.
GalCer-CD1d tetramer+ CD24-TCRβ+ NK1.1- iNKT cells reveals dramatic increase of NKT17 subset.
(b) Percentage of iNKT cells expressing PLZF and Rorγt (NKT17 subset) among the NK1.1− iNKT cells. n=5 WT mice and 7 Tet2-3 DKO mice. Data are compiled from 4 independent experiments.
(c) Representative flow cytometric analysis of PLZF and T-bet expression in NK1.1− iNKT cells. The frequency of PLZF+Tbet+ NKT1 precursor cells is reduced. One representative experiment of 4 is shown.
(d) Percentage of iNKT cells expressing PLZF and T-bet (NKT1 precursor cells) among NK1.1− iNKT cells. p<0.0001. n=5 WT mice and 7 Tet2-3 DKO mice. Data are compiled from 4 independent experiments.
(e) Representative flow cytometric analysis of transcription factors PLZF and T-bet in α-GalCer-CD1d tetramer+ CD24- TCRβ+ NK1.1+ iNKT cells shows expression of RORγt and emergence of an aberrant PLZFhigh Tbet+ population.
(f) α-GalCer-CD1d tetramer+ TCRβ+ NK1.1+ WT and Tet2-3 DKO iNKT cells were stimulated ex vivo with PMA and ionomycin. Representative flow cytometric analysis of α-GalCer-CD1d tetramer+ cells that secrete IFNγ is shown.
(g) Percentage of WT and Tet2-3 DKO α-GalCer-CD1d tetramer+ CD24-TCRβ+NK1.1+ iNKT cells that secrete IFNγ. One of 2 independent experiments is shown. Cells isolated from 3 different mice/ genotype were evaluated.
Data are mean ± SEM. **P< 0.01, ****P< 0.0001 (unpaired t test)
Supplementary Figure 6 Correlation of DNA modifications (5mC and 5hmC) with gene expression
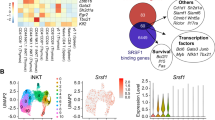
(a) Composite analysis of methylation (5mC+5hmC) in genes with decreased (left, downregulated) or increased (middle, upregulated) expression in young Tet2-3 DKO iNKT cells compared to WT. Right, Methylation in random genomic fragments.
(b) Methylation changes in the promoter (left) and the gene body (right) of individual differentially-expressed genes, plotted against the ratio of their expression in WT versus young Tet2-3 DKO iNKT cells. Each dot represents a gene. Up (red dots) 698 genes with higher expression in WT compared to Tet2-3 DKO iNKT cells; Down (blue dots), 649 genes with lower expression in WT compared to Tet2-3 DKO iNKT cells.
Supplementary Figure 7 Differentially modified regions (DMRs) and differentially accessible regions (DARs) in genes related to the iNKT cell specification program.
Genome browser views of DNA modification (5mC+5hmC) identified by WGBS and chromatin accessibility identified by ATAC-seq in (a) Tbx21; (b), Zbtb7b and (c) Rorc genes in common lymphoid progenitors (CLP, purple), WT iNKT cells (blue) and young Tet2/3 DKO iNKT cells (green). The arrow indicates the direction of transcription. Statistically significant DMRs (black) or DARs (purple for gain of accessibility in Tet2/3 DKO or black for more accessibility in WT iNKT cells) are indicated by horizontal bars. Selected regions in which modification or/and accessibility is altered from CLP to iNKT cells, and then progressively is affected in young iNKT cells are highlighted (salmon shading). For each gene, the gene expression level (RPKM) in each cell type is indicated.
Supplementary Figure 8 Portraits of DNA modification and gene expression of selected downregulated cytokines and transcription factors in Tet2-3 DKO iNKT cells.
(a)-(c), Genome browser views of DNA modification (5mC+5hmC, identified by WGBS) and chromatin accessibility (identified by ATAC-seq) in a, Il4; b, Ifng and c, Bcl11b genes in common lymphoid progenitors (CLP, purple), WT iNKT cells (blue) and young Tet2-3 DKO iNKT cells (green). The arrow indicates the direction of transcription. Statistically significant DMRs (black) or DARs (purple for gain of accessibility in DKO or black for more accessibility in WT) are indicated by horizontal bars. Selected regions in which DNA modification or/and accessibility is altered from CLP to iNKT cells, and then progressively affected in young iNKT cells, are highlighted (salmon shading). For each gene the expression (RPKM) in each cell type is indicated.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–8 and Supplementary Methods (PDF 3474 kb)
Rights and permissions
About this article
Cite this article
Tsagaratou, A., González-Avalos, E., Rautio, S. et al. TET proteins regulate the lineage specification and TCR-mediated expansion of iNKT cells. Nat Immunol 18, 45–53 (2017). https://doi.org/10.1038/ni.3630
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ni.3630
This article is cited by
-
IL-12 reprograms CAR-expressing natural killer T cells to long-lived Th1-polarized cells with potent antitumor activity
Nature Communications (2024)
-
TET (Ten-eleven translocation) family proteins: structure, biological functions and applications
Signal Transduction and Targeted Therapy (2023)
-
Role of TET dioxygenases in the regulation of both normal and pathological hematopoiesis
Journal of Experimental & Clinical Cancer Research (2022)
-
Acute deletion of TET enzymes results in aneuploidy in mouse embryonic stem cells through decreased expression of Khdc3
Nature Communications (2022)
-
CD4 expression in effector T cells depends on DNA demethylation over a developmentally established stimulus-responsive element
Nature Communications (2022)