Abstract
The mammalian cytoplasmic multi-tRNA synthetase complex (MSC) is a depot system that regulates non-translational cellular functions. Here we found that the MSC component glutamyl-prolyl-tRNA synthetase (EPRS) switched its function following viral infection and exhibited potent antiviral activity. Infection-specific phosphorylation of EPRS at Ser990 induced its dissociation from the MSC, after which it was guided to the antiviral signaling pathway, where it interacted with PCBP2, a negative regulator of mitochondrial antiviral signaling protein (MAVS) that is critical for antiviral immunity. This interaction blocked PCBP2-mediated ubiquitination of MAVS and ultimately suppressed viral replication. EPRS-haploid (Eprs+/−) mice showed enhanced viremia and inflammation and delayed viral clearance. This stimulus-inducible activation of MAVS by EPRS suggests an unexpected role for the MSC as a regulator of immune responses to viral infection.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Accession codes
References
Guo, M., Yang, X.L. & Schimmel, P. New functions of aminoacyl-tRNA synthetases beyond translation. Nat. Rev. Mol. Cell Biol. 11, 668–674 (2010).
Ray, P.S., Arif, A. & Fox, P.L. Macromolecular complexes as depots for releasable regulatory proteins. Trends Biochem. Sci. 32, 158–164 (2007).
Sampath, P. et al. Noncanonical function of glutamyl-prolyl-tRNA synthetase: gene-specific silencing of translation. Cell 119, 195–208 (2004).
Mukhopadhyay, R., Jia, J., Arif, A., Ray, P.S. & Fox, P.L. The GAIT system: a gatekeeper of inflammatory gene expression. Trends Biochem. Sci. 34, 324–331 (2009).
Guo, M. & Schimmel, P. Essential nontranslational functions of tRNA synthetases. Nat. Chem. Biol. 9, 145–153 (2013).
Tandle, A.T. et al. Endothelial monocyte activating polypeptide-II modulates endothelial cell responses by degrading hypoxia-inducible factor-1alpha through interaction with PSMA7, a component of the proteasome. Exp. Cell Res. 315, 1850–1859 (2009).
Kim, S., You, S. & Hwang, D. Aminoacyl-tRNA synthetases and tumorigenesis: more than housekeeping. Nat. Rev. Cancer 11, 708–718 (2011).
Kwon, N.H. et al. Dual role of methionyl-tRNA synthetase in the regulation of translation and tumor suppressor activity of aminoacyl-tRNA synthetase-interacting multifunctional protein-3. Proc. Natl. Acad. Sci. USA 108, 19635–19640 (2011).
Ofir-Birin, Y. et al. Structural switch of lysyl-tRNA synthetase between translation and transcription. Mol. Cell 49, 30–42 (2013).
Kim, D.G. et al. Interaction of two translational components, lysyl-tRNA synthetase and p40/37LRP, in plasma membrane promotes laminin-dependent cell migration. FASEB J. 26, 4142–4159 (2012).
Cho, H.Y. et al. Assembly of multi-tRNA synthetase complex via heterotetrameric glutathione transferase-homology domains. J. Biol. Chem. 290, 29313–29328 (2015).
Wolfe, C.L., Warrington, J.A., Treadwell, L. & Norcum, M.T. A three-dimensional working model of the multienzyme complex of aminoacyl-tRNA synthetases based on electron microscopic placements of tRNA and proteins. J. Biol. Chem. 280, 38870–38878 (2005).
Arif, A. et al. Two-site phosphorylation of EPRS coordinates multimodal regulation of noncanonical translational control activity. Mol. Cell 35, 164–180 (2009).
Jia, J., Arif, A., Ray, P.S. & Fox, P.L. WHEP domains direct noncanonical function of glutamyl-prolyl tRNA synthetase in translational control of gene expression. Mol. Cell 29, 679–690 (2008).
Vyas, K. et al. Genome-wide polysome profiling reveals an inflammation-responsive posttranscriptional operon in γ interferon-activated monocytes. Mol. Cell. Biol. 29, 458–470 (2009).
Schoenborn, J.R. & Wilson, C.B. Regulation of interferon-γ during innate and adaptive immune responses. Adv. Immunol. 96, 41–101 (2007).
Schroder, K., Hertzog, P.J., Ravasi, T. & Hume, D.A. Interferon-γ: an overview of signals, mechanisms and functions. J. Leukoc. Biol. 75, 163–189 (2004).
Akira, S., Uematsu, S. & Takeuchi, O. Pathogen recognition and innate immunity. Cell 124, 783–801 (2006).
McWhirter, S.M., Tenoever, B.R. & Maniatis, T. Connecting mitochondria and innate immunity. Cell 122, 645–647 (2005).
Gack, M.U. et al. TRIM25 RING-finger E3 ubiquitin ligase is essential for RIG-I-mediated antiviral activity. Nature 446, 916–920 (2007).
Belgnaoui, S.M. et al. Linear ubiquitination of NEMO negatively regulates the interferon antiviral response through disruption of the MAVS-TRAF3 complex. Cell Host Microbe 12, 211–222 (2012).
You, F. et al. PCBP2 mediates degradation of the adaptor MAVS via the HECT ubiquitin ligase AIP4. Nat. Immunol. 10, 1300–1308 (2009).
Yang, X.L. Structural disorder in expanding the functionome of aminoacyl-tRNA synthetases. Chem. Biol. 20, 1093–1099 (2013).
Sajish, M. et al. Trp-tRNA synthetase bridges DNA-PKcs to PARP-1 to link IFN-γ and p53 signaling. Nat. Chem. Biol. 8, 547–554 (2012).
Arif, A., Jia, J., Moodt, R.A., DiCorleto, P.E. & Fox, P.L. Phosphorylation of glutamyl-prolyl tRNA synthetase by cyclin-dependent kinase 5 dictates transcript-selective translational control. Proc. Natl. Acad. Sci. USA 108, 1415–1420 (2011).
Papatriantafyllou, M. Innate immunity: MAVS build-ups for defence. Nat. Rev. Immunol. 11, 570–571 (2011).
Xia, P. et al. IRTKS negatively regulates antiviral immunity through PCBP2 sumoylation-mediated MAVS degradation. Nat. Commun. 6, 8132 (2015).
Sekine, S. et al. ATP binding by glutamyl-tRNA synthetase is switched to the productive mode by tRNA binding. EMBO J. 22, 676–688 (2003).
Son, J. et al. Conformational changes in human prolyl-tRNA synthetase upon binding of the substrates proline and ATP and the inhibitor halofuginone. Acta Crystallogr. D Biol. Crystallogr. 69, 2136–2145 (2013).
van den Berg, A. & Dowdy, S.F. Protein transduction domain delivery of therapeutic macromolecules. Curr. Opin. Biotechnol. 22, 888–893 (2011).
Cahuzac, B., Berthonneau, E., Birlirakis, N., Guittet, E. & Mirande, M. A recurrent RNA-binding domain is appended to eukaryotic aminoacyl-tRNA synthetases. EMBO J. 19, 445–452 (2000).
Zhou, H., Sun, L., Yang, X.L. & Schimmel, P. ATP-directed capture of bioactive herbal-based medicine on human tRNA synthetase. Nature 494, 121–124 (2013).
Rubin, S.M. Deciphering the retinoblastoma protein phosphorylation code. Trends Biochem. Sci. 38, 12–19 (2013).
Burkhart, D.L. & Sage, J. Cellular mechanisms of tumour suppression by the retinoblastoma gene. Nat. Rev. Cancer 8, 671–682 (2008).
Yoneyama, M. et al. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat. Immunol. 5, 730–737 (2004).
Takeuchi, O. & Akira, S. Pattern recognition receptors and inflammation. Cell 140, 805–820 (2010).
Wang, Y., Tong, X. & Ye, X. Ndfip1 negatively regulates RIG-I-dependent immune signaling by enhancing E3 ligase Smurf1-mediated MAVS degradation. J. Immunol. 189, 5304–5313 (2012).
Jacobs, J.L., Zhu, J., Sarkar, S.N. & Coyne, C.B. Regulation of mitochondrial antiviral signaling (MAVS) expression and signaling by the mitochondria-associated endoplasmic reticulum membrane (MAM) protein Gp78. J. Biol. Chem. 289, 1604–1616 (2014).
Castanier, C. et al. MAVS ubiquitination by the E3 ligase TRIM25 and degradation by the proteasome is involved in type I interferon production after activation of the antiviral RIG-I-like receptors. BMC Biol. 10, 44 (2012).
Rosen, B., Schick, J. & Wurst, W. Beyond knockouts: the International Knockout Mouse Consortium delivers modular and evolving tools for investigating mammalian genes. Mamm. Genome 26, 456–466 (2015).
Pascua, P.N. et al. Virulence and transmissibility of H1N2 influenza virus in ferrets imply the continuing threat of triple-reassortant swine viruses. Proc. Natl. Acad. Sci. USA 109, 15900–15905 (2012).
Kim, S.K. et al. A nineteen gene-based risk score classifier predicts prognosis of colorectal cancer patients. Mol. Oncol. 8, 1653–1666 (2014).
Han, J.M. et al. Leucyl-tRNA synthetase is an intracellular leucine sensor for the mTORC1-signaling pathway. Cell 149, 410–424 (2012).
French, A.P., Mills, S., Swarup, R., Bennett, M.J. & Pridmore, T.P. Colocalization of fluorescent markers in confocal microscope images of plant cells. Nat. Protoc. 3, 619–628 (2008).
Kim, K. et al. Reinvestigation of aminoacyl-tRNA synthetase core complex by affinity purification-mass spectrometry reveals TARSL2 as a potential member of the complex. PLoS One 8, e81734 (2013).
Acknowledgements
Supported by the KRIBB Initiative Program (KGM4541622 to M.H.K.), the National Research Foundation of Korea, funded by the Ministry of Science, ICT & Future Planning of Korea (NRF-2010-0029767 and 2014R1A2A1A01005971 to M.H.K.; NRF-M3A6A4-2010-0029785 to S.K.; and 2015020957 to J.-S.L.), the Korea Institute of Oriental Medicine (K12050 to J.-S.L.), the Ministry for Food, Agriculture, Forestry and Fisheries (315044031SB010 to J.-S.L.) and the Korea Health Industry Development Institute (HI14C3484 to C.L.).
Author information
Authors and Affiliations
Contributions
E.-Y.L. and H.-C.L. performed most of the experiments with help from H.-K.K., S.Y.J., J.H., J.-H.K. and T.-H.K. S.-J.P. and C.L. performed mass spectrometry. Y.-H.K. and C.-H.L. performed immunohistochemical analysis. J.H.K., S.-Y.K. and Y.-K.C. performed RNA-seq analysis. A.A., J.U.J., P.L.F. and S.K. contributed to the discussion and provided critical reagents. E.-Y.L., J.-S.L. and M.H.K. designed the study and wrote the manuscript. All of the authors helped with data analysis.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
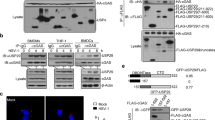
Supplementary Figure 1 EPRS expression in multiple cell lines upon viral infection.
(a,b) EPRS is slightly induced upon viral induction. qPCR of Eprs mRNA (a) and immunoblot analysis of corresponding endogenous EPRS expression (b) in multiple cell lines, including C57/B6 mouse-derived BMDM, U937, RAW264.7, A549, and 293T cells infected with PR8-GFP or VSV-GFP. (c) qPCR analysis of representative Isg mRNA expression under the same conditions as in (a). (d) qPCR analysis of Eprs mRNA in U937 and RAW264.7 cells treated with IFN-β (1000 units/ml). Isg15 mRNA was analyzed as a control. (e) qPCR analysis of Eprs mRNA in RIG-I-sufficient (Ddx58+/+) or RIG-I-deficient (Ddx58-/-) MEF cells infected with VSV-GFP. Data are representative of two (a-e) independent biological replicates with similar results (mean and s.d. of triplicate in a,c-e).
Supplementary Figure 2 Antiviral effects of EPRS in EPRS-deficient or EPRS-overexpressing immune cells.
(a) Immunoblot analysis of EPRS expression. (b) Viral titer after infection with HSV-GFP (MOI = 1). RAW264.7 cells were transfected with non-targeting control siRNA (siCtrl) or siEPRS (a,b). (c) Fluorescence microscopy images, (d) virus replication, and (e) secreted IFN-β or IL-6 levels in 293T cells transfected with siCtrl or siEPRS for 36 h, followed by infection with VSV-GFP (MOI = 0.0001). (f) Immunoblot analysis of EPRS expression. (g) Fluorescence microscopy images, (h) PR8 titer, and (i) secreted IFN-β or IL-6 level in cells infected with PR8-GFP (MOI = 1). (j) Fluorescence microscopy images, (k) VSV titer, and (l) secreted IFN-β or IL-6 level in stable EPRS-deficient cells infected with VSV-GFP (MOI = 0.5). RAW264.7 cells were transduced with non-targeting control shRNA (shCtrl) or EPRS shRNA (shEPRS), followed by selection with puromycin (f–l). (m) Immunoblot analysis of EPRS expression. (n) Fluorescence microscopy images, (o) VSV titers, and (p) secreted IFN-β or IL-6 level in EPRS-overexpressing cells infected with VSV-GFP (MOI = 0.5). RAW264.7 cells were transfected with a FLAG-tagged empty vector (Ctrl) or with EPRS-FLAG (EPRS) plasmids, followed by selection with puromycin (m–p). Scale bars, 100 μm (c,g,j,n). *P < 0.05, **P < 0.01, and ***P < 0.001 (Student’s t-test; d,e,h,i,k,l,o,p). Data are representative of three (a-p) independent biological replicates with similar results (mean and s.d. of triplicate in b,d,e,h,i,k,l,o,p).
Supplementary Figure 3 EPRS deficiency in mouse BMDMs reduces antiviral innate immune responses.
(a) Immunoblot analysis of EPRS expression in BMDMs. BMDMs were transfected with non-targeting control siRNA (siCtrl) or siEPRS for 36 h. (b) Plaque assay to determine virus titers and (c) ELISA to measure IFN-β and IL-6 levels at 12 and 24 h post-infection. BMDMs were infected with PR8-GFP (MOI = 3) or VSV-GFP (MOI = 5) (b,c). (d) IFN-β and IL-6 levels measured in the culture supernatants from BMDMs treated with 40 μg of Poly(I:C). (e) Induction of Ifnb mRNA or IFN-related antiviral genes in virus-infected cells. RAW264.7 cells were transfected with siCtrl or siEPRS for 36 h, followed by infection with PR8-GFP (MOI = 1) for 12 h. The graphs show the -fold induction of the indicated genes after normalization against Gapdh. (f) Viral titer (determined by plaque assay) and (g) secreted IFN-β or IL-6 levels in cell culture supernatants after infection with HSV-GFP. BMDMs from Eprs+/+ and Eprs+/ – mice were infected with HSV-GFP (MOI = 2) (f,g). *P < 0.05, **P < 0.01, and ***P < 0.001 (Student’s t-test; b–d). Data are representative of three (b-d) or two (e-g) independent biological replicates with similar results (mean and s.d. of triplicate in b-g).
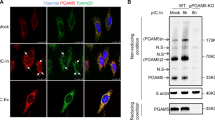
Supplementary Figure 4 Dissociation of EPRS from the MSC upon viral infection.
(a,b) Viral infection induces dissociation of EPRS from the MSC component proteins. Lysates of RAW264.7 cells infected with PR8-GFP (MOI = 1) were subjected to immunoprecipitation with an anti-EPRS (a) or with an anti-KRS (b), followed by immunoblot analysis with anti-KRS, anti-MRS, anti-AIMP3, and anti-GAPDH (a), or with anti-EPRS and anti-AIMP3 (b), respectively. (c) Confocal microscopy of endogenous EPRS (red) and KRS (green) in HeLa cells infected with PR8 (MOI = 5) for 6 or 12 h. IFN-γ (1000 units/ml) treatment for 12 h was used for comparison. Cells were permeabilized with a lower dose of digitonin (20 μg/ml, 5 min) than used in Fig. 4b (25 μg/ml, 10 min). Scale bar, 10 μm (2 μm in magnified images). (d) Confocal microscopy of endogenous EPRS (red) and NSAP1 (green) in HeLa cells infected with PR8 (MOI = 5) for 6 or 12 h, or in cells treated with IFN-γ (1000 units/ml) for 12 h. Scale bar, 10 μm. Data are representative of three independent biological replicates with similar results (a-d).
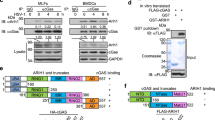
Supplementary Figure 5 Identification of the viral-infection-specific phosphorylation site in EPRS.
(a) Silver-stained Strep-EPRS (indicated by an asterisk) purified by Strep precipitation assay of 293T cells infected or uninfected (–) with PR8-GFP (MOI = 5). EV, Strep-empty vector. (b) MS/MS spectra for a doubly charged EPRS peptide EYIPGQPPLSQSSDSpS*PTR (MH+ = 2125.93, z = 2+) obtained under uninfected (upper) and PR8-infected (lower panel) conditions. The peptides contain the S886 phosphorylation site (marked by an asterisk). (c–e) Extracted ion chromatogram (XIC) of the tryptic digests under uninfected (upper) and infected (lower panel) conditions, corresponding to doubly charged EYIPGQPPLSQSSDSSPTR (MH+ = 2044.96, z = 2+) (c), doubly charged NQGGGLSSSGAGEGQGPK (MH+ = 1586.72, z = 2+) (d), and triply charged KDPSKNQGGGLSSSGAGEGQGPK (MH+ = 2142.02, z = 3+) (e) peptides from non-phosphorylated (left) and phosphorylated (right, marked by an asterisk) EPRS. ND, not detected. (f) Immunoblot analysis of phosphomimetic (S990D) and phosphorylation-resistant (S990A) EPRS against with anti-phospho-EPRS(Ser990) in 293T cells. (g–j) Immunoblot analysis of EPRS Ser990 phosphorylation in RAW264.7 cells infected with PR8-GFP (MOI =1) (g), 293T cells infected with PR8-GFP (MOI = 5) (h) or VSV-GFP (MOI = 0.001) (i), or cells transfected with 2 μg of Poly(I:C) (j). (k,l) Secreted IFN-γ levels from U937 (k) or RAW264.7 (l) cells infected with PR8-GFP or VSV-GFP. Cells treated with IFN-γ (1000 units/ml) were used as a positive control. (m) Immunoblot analysis of Cp expression in PR8-GFP-infected RAW264.7 cells. Data are representative of three (g-m) independent biological replicates with similar results (mean and s.d. of triplicate in k,l).
Supplementary Figure 6 Non-translational role of EPRS in regulating antiviral immune responses.
(a,b) Purified His-tagged EPRS (aa 1–196) (a) or EPRS (aa 1–168) (b) was mixed with the GST-fused PCBP2 KH1 (aa 11-82). After His-tag precipitation, proteins were subjected to SDS-PAGE and stained with Coomassie Brilliant Blue. (c) The purified His-tagged ERS (aa 1-732) and its mutant that is inactive for tRNA glutamylation (MT). Black arrows denote protein fragments derived from ERS during purification (as in Fig. 6j). (d) Aminoacylation assay for ERS (WT) and its mutant (MT). CPM, counts per minute. Ctrl, buffer without protein. (e) IFNB promoter activity in 293T cells transfected with N-RIG-I plus empty vector (EV), Strep-EPRS (WT), or its mutants inactive for tRNA glutamylation only (E-MT), tRNA prolylation only (P-MT), or both (EP-MT). (f) Immunoblot analysis of endogenous EPRS, MAVS, and RIG-I expression in sgEPRS 293T cells or non-targeting control (sgCtrl) cells. (g–k) Non-translational function of EPRS in antiviral immune responses. Virus replication assay (examined by fluorescence microscopy) (g) and plaque assay (h) at 24 h post-infection with VSV-GFP (MOI = 0.0001). Immunoblot analysis of Strep-EPRS or endogenous EPRS expression (i). IFN-β (j) and IL-6 (k) secreted by sgEPRS cells infected with VSV-GFP. sgCtrl or sgEPRS 293T cells were reconstituted with EV, Strep-tagged EPRS (WT), or its catalytic mutant (EP-MT) (g–k). Scale bar, 100 μm (g). *P < 0.01; NS, not significant (Student’s t-test; d,e,h,j,k). Data are representative of two (a-k) independent experiments (mean and s.d. of triplicate in d,e,h,j,k).
Supplementary Figure 7 Infection-specific EPRS phosphorylation is essential for regulating MAVS.
(a,b) In vitro binding assay showing MAVS interaction with PCBP2 KH1 (aa 11–82). (c,d) The precipitation (ppt) assays revealed no interaction between PCBP2 and LRS in 293T cells (c), whereas PCBP2 interacted with MAVS (d). (e) Ubiquitination of exogenous MAVS in non-targeting control (siCtrl) or EPRS-deficient (siEPRS) 293T cells transfected with Ub, ITCH, MAVS, or PCBP2. (f) Ubiquitination of endogenous MAVS in 293T cells transfected with Ub, ITCH, PCBP2, and Strep-empty vector (EV), or with WT EPRS or its mutant (EP-MT, enzymatically inactive for both tRNA glutamylation and prolylation). (g) Expression of endogenous MAVS in 293T cells transfected with PCBP2 and WT EPRS or EP-MT. The histogram shows the intensity of the MAVS band normalized against actin. (h–n) Ubiquitination of endogenous MAVS (h) in non-targeting control (sgCtrl) or sgEPRS 293T cells transfected with HA-Ub and infected with VSV-GFP (MOI = 0.1). (i) Ubiquitination of endogenous MAVS in 293T cells transfected with HA-Ub and infected with VSV-GFP. (j) IFN-β or (k) IL-6 levels in supernatants from cells infected with VSV-GFP. (l) Fluorescence microscopy images and (m) plaque assay at 24 h post-infection with VSV-GFP (MOI = 0.0001). (n) Immunoblot analysis of EPRS-FLAG or endogenous EPRS. sgEPRS 293T cells were reconstituted with a FLAG-EV, WT EPRS, S990A, or S990D (i–n). Scale bar, 100 μm (l). *P < 0.01; NS, not significant (Student’s t-test; j,k,m). Data are representative of two (a-n) independent biological replicates with similar results (mean and s.d. of triplicate in j,k,m).
Supplementary Figure 8 The Tat-Epep is specific to infection with RNA viruses.
(a–c) Tat-Epep has no significant effect on virus replication (a) or IFN-β (b) and IL-6 (c) secretion in RAW264.7 infected with HSV-GFP (MOI = 1) for 12 h. HSV-GFP-infected RAW264.7 cells treated with PBS were used as negative controls. (d) Viability of RAW264.7 cells as measured in an MTS assay after treatment with the indicated doses of Tat-Epep for 12 h. (e) Viability of 293T cells after treatment with Tat-Epep for 12 or 24 h. Ctrl, 293T cells treated with a lytic detergent (digitonin, 30 μg/ml) for 15 min as a positive control. Results are expressed as the mean ± SD of two independent biological replicates incorporating triplicate samples.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–8 and Supplementary Tables 1 and 2 (PDF 1905 kb)
Rights and permissions
About this article
Cite this article
Lee, EY., Lee, HC., Kim, HK. et al. Infection-specific phosphorylation of glutamyl-prolyl tRNA synthetase induces antiviral immunity. Nat Immunol 17, 1252–1262 (2016). https://doi.org/10.1038/ni.3542
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ni.3542
This article is cited by
-
ChemRAP uncovers specific mRNA translation regulation via RNA 5′ phospho-methylation
EMBO Reports (2024)
-
A viral pan-end RNA element and host complex define a SARS-CoV-2 regulon
Nature Communications (2023)
-
Immune inhibitory receptor-mediated immune response, metabolic adaptation, and clinical characterization in patients with COVID-19
Scientific Reports (2023)
-
Glutamyl-prolyl-tRNA synthetase 1 coordinates early endosomal anti-inflammatory AKT signaling
Nature Communications (2022)
-
Meta-omics characteristics of intestinal microbiota associated to HBeAg seroconversion induced by oral antiviral therapy
Scientific Reports (2021)