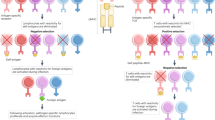
Abstract
The cells that stimulate positive selection express specialized proteasome β-subunits different from those expressed by all other cells, including those involved in negative selection. Mice that lack all four specialized proteasome β-subunits, and therefore express only constitutive proteasomes in all cells, had a profound defect in the generation of CD8+ T cells. While a defect in positive selection would reflect an inability to generate the appropriate positively selecting peptides, a block at negative selection would point to the potential need to switch peptides between positive selection and negative selection to avoid the two processes' often cancelling each other out. We found that the block in T cell development occurred around the checkpoints of positive selection and, unexpectedly, negative selection as well.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Starr, T.K., Jameson, S.C. & Hogquist, K.A. Positive and negative selection of T cells. Annu. Rev. Immunol. 21, 139–176 (2003).
Singer, A., Adoro, S. & Park, J.-H. Lineage fate and intense debate: myths, models and mechanisms of CD4- versus CD8-lineage choice. Nat. Rev. Immunol. 8, 788–801 (2008).
Rock, K.L. et al. Inhibitors of the proteasome block the degradation of most cell proteins and the generation of peptides presented on MHC class I molecules. Cell 78, 761–771 (1994).
Coux, O., Tanaka, K. & Goldberg, A.L. Structure and functions of the 20S and 26S proteasomes. Annu. Rev. Biochem. 65, 801–847 (1996).
Murata, S. et al. Regulation of CD8+ T cell development by thymus-specific proteasomes. Science 316, 1349–1353 (2007).
Guillaume, B. et al. Two abundant proteasome subtypes that uniquely process some antigens presented by HLA class I molecules. Proc. Natl. Acad. Sci. USA 107, 18599–18604 (2010).
Kisselev, A.F., Akopian, T.N., Castillo, V. & Goldberg, A.L. Proteasome active sites allosterically regulate each other, suggesting a cyclical bite-chew mechanism for protein breakdown. Mol. Cell 4, 395–402 (1999).
Kincaid, E.Z. et al. Mice completely lacking immunoproteasomes show major changes in antigen presentation. Nat. Immunol. 13, 129–135 (2012).
Sasaki, K. et al. Thymoproteasomes produce unique peptide motifs for positive selection of CD8+ T cells. Nat. Commun. 6, 7484 (2015).
Xing, Y., Jameson, S.C. & Hogquist, K.A. Thymoproteasome subunit-β5T generates peptide-MHC complexes specialized for positive selection. Proc. Natl. Acad. Sci. USA 110, 6979–6984 (2013).
Hogquist, K.A., Jameson, S.C. & Bevan, M.J. Strong agonist ligands for the T cell receptor do not mediate positive selection of functional CD8+ T cells. Immunity 3, 79–86 (1995).
Klein, L., Kyewski, B., Allen, P.M. & Hogquist, K.A. Positive and negative selection of the T cell repertoire: what thymocytes see (and don't see). Nat. Rev. Immunol. 14, 377–391 (2014).
Nitta, T. et al. Thymoproteasome shapes immunocompetent repertoire of CD8+ T cells. Immunity 32, 29–40 (2010).
Klein, L., Hinterberger, M., Wirnsberger, G. & Kyewski, B. Antigen presentation in the thymus for positive selection and central tolerance induction. Nat. Rev. Immunol. 9, 833–844 (2009).
Azzam, H.S. et al. CD5 expression is developmentally regulated by T cell receptor (TCR) signals and TCR avidity. J. Exp. Med. 188, 2301–2311 (1998).
Takada, K. et al. TCR affinity for thymoproteasome-dependent positively selecting peptides conditions antigen responsiveness in CD8+ T cells. Nat. Immunol. 16, 1069–1076 (2015).
Hogquist, K.A. et al. Identification of a naturally occurring ligand for thymic positive selection. Immunity 6, 389–399 (1997).
Yamashita, I., Nagata, T., Tada, T. & Nakayama, T. CD69 cell surface expression identifies developing thymocytes which audition for T cell antigen receptor-mediated positive selection. Int. Immunol. 5, 1139–1150 (1993).
Davey, G.M. et al. Preselection thymocytes are more sensitive to T cell receptor stimulation than mature T cells. J. Exp. Med. 188, 1867–1874 (1998).
Egawa, T. & Littman, D.R. ThPOK acts late in specification of the helper T cell lineage and suppresses Runx-mediated commitment to the cytotoxic T cell lineage. Nat. Immunol. 9, 1131–1139 (2008).
Grueter, B. et al. Runx3 regulates integrin alpha E/CD103 and CD4 expression during development of CD4−/CD8+ T cells. J. Immunol. 175, 1694–1705 (2005).
Takeuchi, Y., Habu, S., Okumura, K. & Suzuki, G. Cyclosporin A and anti-Ia antibody cause a maturation defect of CD4+8− cells in organ-cultured fetal thymus. Immunology 66, 362–367 (1989).
Ueno, T. et al. CCR7 signals are essential for cortex-medulla migration of developing thymocytes. J. Exp. Med. 200, 493–505 (2004).
Egawa, T., Tillman, R.E., Naoe, Y., Taniuchi, I. & Littman, D.R. The role of the Runx transcription factors in thymocyte differentiation and in homeostasis of naive T cells. J. Exp. Med. 204, 1945–1957 (2007).
Adoro, S. et al. Coreceptor gene imprinting governs thymocyte lineage fate. EMBO J. 31, 366–377 (2012).
Merkenschlager, M. et al. Centromeric repositioning of coreceptor loci predicts their stable silencing and the CD4/CD8 lineage choice. J. Exp. Med. 200, 1437–1444 (2004).
Stritesky, G.L. et al. Murine thymic selection quantified using a unique method to capture deleted T cells. Proc. Natl. Acad. Sci. USA 110, 4679–4684 (2013).
Takahama, Y. Journey through the thymus: stromal guides for T-cell development and selection. Nat. Rev. Immunol. 6, 127–135 (2006).
Bouillet, P. et al. BH3-only Bcl-2 family member Bim is required for apoptosis of autoreactive thymocytes. Nature 415, 922–926 (2002).
Suen, A.Y.W. & Baldwin, T.A. Proapoptotic protein Bim is differentially required during thymic clonal deletion to ubiquitous versus tissue-restricted antigens. Proc. Natl. Acad. Sci. USA 109, 893–898 (2012).
Gray, D.H.D. et al. The BH3-only proteins Bim and Puma cooperate to impose deletional tolerance of organ-specific antigens. Immunity 37, 451–462 (2012).
Lehmann-Grube, F., Dralle, H., Utermöhlen, O. & Löhler, J. MHC class I molecule-restricted presentation of viral antigen in β2-microglobulin-deficient mice. J. Immunol. 153, 595–603 (1994).
Dolan, B.P., Gibbs, K.D. Jr. & Ostrand-Rosenberg, S. Dendritic cells cross-dressed with peptide MHC class I complexes prime CD8+ T cells. J. Immunol. 177, 6018–6024 (2006).
McCaughtry, T.M., Baldwin, T.A., Wilken, M.S. & Hogquist, K.A. Clonal deletion of thymocytes can occur in the cortex with no involvement of the medulla. J. Exp. Med. 205, 2575–2584 (2008).
Marrack, P. & Kappler, J. The T cell receptor. Science 238, 1073–1079 (1987).
Derbinski, J. et al. Promiscuous gene expression in thymic epithelial cells is regulated at multiple levels. J. Exp. Med. 202, 33–45 (2005).
Honey, K. & Rudensky, A.Y. Lysosomal cysteine proteases regulate antigen presentation. Nat. Rev. Immunol. 3, 472–482 (2003).
Gommeaux, J. et al. Thymus-specific serine protease regulates positive selection of a subset of CD4+ thymocytes. Eur. J. Immunol. 39, 956–964 (2009).
Nussbaum, A.K., Rodriguez-Carreno, M.P., Benning, N., Botten, J. & Whitton, J.L. Immunoproteasome-deficient mice mount largely normal CD8+ T cell responses to lymphocytic choriomeningitis virus infection and DNA vaccination. J. Immunol. 175, 1153–1160 (2005).
Mandl, J.N., Monteiro, J.P., Vrisekoop, N. & Germain, R.N. T cell-positive selection uses self-ligand binding strength to optimize repertoire recognition of foreign antigens. Immunity 38, 263–274 (2013).
Baldwin, T.A., Hogquist, K.A. & Jameson, S.C. The fourth way? Harnessing aggressive tendencies in the thymus. J. Immunol. 173, 6515–6520 (2004).
Kurobe, H. et al. CCR7-dependent cortex-to-medulla migration of positively selected thymocytes is essential for establishing central tolerance. Immunity 24, 165–177 (2006).
Anderson, M.S. et al. Projection of an immunological self shadow within the thymus by the aire protein. Science 298, 1395–1401 (2002).
Reiser, H. & Schneeberger, E.E. The costimulatory molecule B7 is expressed in the medullary region of the murine thymus. Immunology 81, 532–537 (1994).
Jenkinson, E.J., Anderson, G., Moore, N.C., Smith, C.A. & Owen, J.J. Positive selection by purified MHC class II+ thymic epithelial cells in vitro: costimulatory signals mediated by B7 are not involved. Dev. Immunol. 3, 265–271 (1994).
Sprent, J. & Surh, C.D. Re-entry of mature T cells to the thymus: an epiphenomenon? Immunol. Cell Biol. 87, 46–49 (2009).
Ernst, B., Lee, D.S., Chang, J.M., Sprent, J. & Surh, C.D. The peptide ligands mediating positive selection in the thymus control T cell survival and homeostatic proliferation in the periphery. Immunity 11, 173–181 (1999).
Goldrath, A.W. & Bevan, M.J. Low-affinity ligands for the TCR drive proliferation of mature CD8+ T cells in lymphopenic hosts. Immunity 11, 183–190 (1999).
Ohta, Y., McKinney, E.C., Criscitiello, M.F. & Flajnik, M.F. Proteasome, transporter associated with antigen processing, and class I genes in the nurse shark Ginglymostoma cirratum: evidence for a stable class I region and MHC haplotype lineages. J. Immunol. 168, 771–781 (2002).
Ramsdell, F. in Current Protocols in Immunology (eds. Coligan, J.E., Bierer, B.E., Margulies, D.H., Shevach, E.M. & Strober, W.) (John Wiley & Sons, 2001).
Acknowledgements
We thank D. Littman (New York University School of Medicine) for Runx3dYFP/YFP mice, and A. Singer and E. Huseby for discussions. Core resources of the University of Massachusetts were used (supported by the Diabetes Endocrinology Research Center (DK32520)). Supported by the US National Institutes of Health (AI20248 and AI110374 to K.L.R., and T32CA130807-02 to E.Z.K.), the University of Massachusetts Diabetes and Endocrine Research Center (DK32520 for K.R.) and Japan Society for the Promotion of Science KAKENHI (21000012 to K.T., and 25221102 to S.M.).
Author information
Authors and Affiliations
Contributions
E.Z.K. designed and did experiments, analyzed data and wrote the paper; S.M. and K.T. generated the β5t-defidient mice and discussed results and conclusions; and K.L.R. designed experiments, supervised the experiments and wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
Supplementary Figure 1 Additional staining and gating of 4KO thymi.
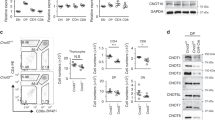
(a) Quantitation of total thymocytes in wild-type and 4KO animals. (n = 8 mice per genotype, male and female) (b) Flow cytometry of WT and 4KO thymi. Numbers in quadrants indicate percent CD4+CD8α+ (uppser left) or CD4−CD8α− (upper right) cells among total thymocytes. (c,d) Quantitation of CD4−CD8α− (c) or CD4+CD8α+ (d) thymocytes in wild-type and 4KO mice, as in b. (e) Quantitation of CD19+B220+ cells in wild-type thymi, wild-type inguinal lymph nodes (LN), 4KO thymi, or 4KO inguinal lymph nodes. (n = 6 mice per genotype, male and female) (f) Quantitation of CD4−CD8α+TCRβhi and CD4−CD8β+TCRβhi thymocytes in wild-type (n = 9, male and female) and 4KO (n = 8, male and female) mice. (g) Flow cytometry of CD8α vs CD8β on CD4−CD8α+TCRβhi thymocytes in wild-type and 4KO mice. (h,i) Geometric mean fluorescence intensity of CD5 on CD4−CD8+TCRβhi (h) and CD4+CD8−TCRβhi (i) thymocytes. Each symbol (a,c,d,e,f,h,i) represents an individual mouse; small horizontal lines indicate the mean (± s.d.). * P=0.0079 ** P<0.0001 (Student’s t-test). Data are representative of two experiments (b,g) or are pooled from two experiments (a,c,d,e,f,h,i)
Supplementary Figure 2 TCR Vβ analysis.
(a,b) Frequency of cells staining positive for each indicated Vβ among CD4+ (a) or CD8+ (b) splenocytes. (n = 8 mice per genotype, male and female). Bars indicate the mean (± s.d) * P=0.0101 ** P=0.0019 *** P=0.0008 **** P=0.0006 ***** P<0.0001 (Student’s t-test). There is no significant difference between wild-type and 4KO by Two-way ANOVA. Data are pooled from two experiments.
Supplementary Figure 3 Normal development of CD8+ T cells in mice heterozygous for deficiency in H2-Kb and H2-Db despite a decrease in MHC class I on cTECs similar to that in 4KO mice.
(a) Geometric mean fluorescence intensity of H2-Kb on wild-type (n=8, male and female), H2-Kb+/ –H2-Db+/– (n=4, male and female), and 4KO (n=4, male and female) splenocytes. (b) Quantitation of CD4−CD8+TCRβhi thymocytes in wild-type and H2-Kb+/ –H2-Db+/– mice. (c) Flow cytometry of wild-type CD45−CD326+ adherent thymocytes. Numbers in quadrants indicate percent CD249+UEA-1− (cTEC)(upper right) or CD249−UEA-1+ (mTEC)(lower left) cells among CD45−CD326+ adherent thymocytes. (d,e) Geometric mean fluorescence intensity of H2-Kb on wild-type (n=6, male and female), H2-Kb+/ –H2-Db+/– (n=3, male and female), and 4KO (n=5, male and female) mice on CD45−CD249+CD326+ (d) or CD45−CD249+CD326+ (e) cells, presented relative to that in wild-type mice, set as 100%. Each symbol (a,b,d,e) represents an individual mouse; small horizontal lines indicate the mean (± s.d.). * P<0.05 (One-way ANOVA with Dunnett’s multiple comparison post-test). Data are representative of three experiments (c), or are pooled from four (a,b) or three (d,e) experiments.
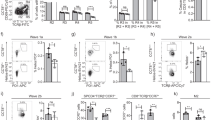
Supplementary Figure 4 4KO thymocytes receive positive selection signals independently of MHC class II.
(a) Quantification of CD69+TCRβint thymocytes in wild-type and 4KO fetal thymic organ cultures in the presence of monoclonal antibody to MHC class II (500 μg/ml). (b) Quantification of wild-type and 4KO CD4+CD8−TCRβhi thymocytes in the presence of control monoclonal antibody (LTF-2) or monoclonal antibody to MHC class II (M5/114) (each at a concentration of 500 μg/ml). (n = 20 lobes (WT + LTF-2), n = 21 lobes (WT + M5/114), n = 21 lobes (4KO + LTF-2) or n = 17 lobes (4KO + M5/114)). Each symbol represents an individual mouse; small horizontal lines indicate the mean (± s.d.). * P=0.0058 and ** P=0.0012 (Student’s t-test). Data are pooled from five experiments.
Supplementary Figure 5 Alternative gating of wild-type and 4KO thymocytes with CD69 and TCRβ.
(a) Flow cytometry of wild-type and 4KO thymocytes (n = 9 mice per genotype, male and female). Numbers in outlined areas indicate percent CD69−TCRβ −cells (bottom left), CD69+TCRβint cells (top middle), CD69+TCRβ hi cells (top right), or CD69− TCRβ hi cells (bottom right) among total thymocytes. Numbers in quadrants indicate percent of cells in that quadrant and the various populations (above plots) gated as on the far left. (b,c,d) Quantitation of CD4+CD8+(b), CD4+CD8− (c) and CD4−CD8+ (d) thymocytes in wild-type (open bars) and 4KO (closed bars) mice gated as in a. Bars indicate the mean (± s.d) Data are representative of three experiments (a), or are pooled from three (b,c,d) experiments.
Supplementary Figure 6 Effects of Bim deficiency on CD4+CD8+ double-positive and CD4SP thymocytes are similar in wild-type and 4KO thymi.
(a,b) Quantitation of CD4+CD8+(a) and CD4+CD8-TCRβhi (b) thymocytes in WT→WT (n = 22), Bcl2l11−/− →WT (n = 16), WT→4KO (n = 16), or Bcl2l11−/− →4KO (n = 16) chimeras. (c) Fold increase in CD4+CD8−TCRβhi thymocytes in Bcl211−/−→WT vs WT→WT chimeras (WT), and Bcl211−/−→4KO vs. WT→4KO chimeras (4KO). (Flow cytometry shown in Fig 4) Each symbol represents an individual mouse; small horizontal lines indicate the mean (± s.d.). * P=0.0289 (Student’s t-test). Data are pooled from 5 experiments.
Supplementary Figure 7 Peptide switching allows many thymocytes to survive negative selection.
In wild-type animals, thymocytes are positively selected on peptides produced by thymoproteasomes in cTECs. Many positively selected thymocytes subsequently avoid deletion because they are negatively selected on largely different peptides, produced by constitutive, immuno or mixed proteasomes in mTECs and DCs, and consequently don’t encounter their high affinity ligands. In the 4KO mice, there is no peptide switching, and most CD8 lineage cells die during negative selection.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–7 (PDF 956 kb)
Supplementary Table 1
Flow cytometry antibody list (XLSX 40 kb)
Rights and permissions
About this article
Cite this article
Kincaid, E., Murata, S., Tanaka, K. et al. Specialized proteasome subunits have an essential role in the thymic selection of CD8+ T cells. Nat Immunol 17, 938–945 (2016). https://doi.org/10.1038/ni.3480
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ni.3480
This article is cited by
-
Arg-tRNA synthetase links inflammatory metabolism to RNA splicing and nuclear trafficking via SRRM2
Nature Cell Biology (2023)
-
Heterozygous missense variant of the proteasome subunit β-type 9 causes neonatal-onset autoinflammation and immunodeficiency
Nature Communications (2021)
-
On the role of the immunoproteasome in transplant rejection
Immunogenetics (2019)
-
Thymoproteasome and peptidic self
Immunogenetics (2019)
-
The immunoproteasome and thymoproteasome: functions, evolution and human disease
Nature Immunology (2018)