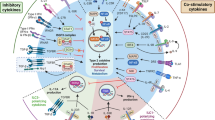
Abstract
Innate lymphoid cells (ILCs) are critical mediators of mucosal immunity, and group 1 ILCs (ILC1 cells) and group 3 ILCs (ILC3 cells) have been shown to be functionally plastic. Here we found that group 2 ILCs (ILC2 cells) also exhibited phenotypic plasticity in response to infectious or noxious agents, characterized by substantially lower expression of the transcription factor GATA-3 and a concomitant switch to being ILC1 cells that produced interferon-γ (IFN-γ). Interleukin 12 (IL-12) and IL-18 regulated this conversion, and during viral infection, ILC2 cells clustered within inflamed areas and acquired an ILC1-like phenotype. Mechanistically, these ILC1 cells augmented virus-induced inflammation in a manner dependent on the transcription factor T-bet. Notably, IL-12 converted human ILC2 cells into ILC1 cells, and the frequency of ILC1 cells in patients with chronic obstructive pulmonary disease (COPD) correlated with disease severity and susceptibility to exacerbations. Thus, functional plasticity of ILC2 cells exacerbates anti-viral immunity, which may have adverse consequences in respiratory diseases such as COPD.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Change history
05 May 2016
In the version of this article initially published online, the units in the vertical axis for Figure 1l were incorrectly stated as '(fold)', and the figure lacked a key. The correct units are '(ng/ml; change from mock)'; the white bars are IL-5, the gray bars are IL-13, and the black bars are IFN-γ. Also, the lowest portions of the images in Figure 4i,j (including the inset in Figure 4j) were incorrectly cropped. The errors have been corrected for the print, PDF and HTML versions of this article.
References
Artis, D. & Spits, H. The biology of innate lymphoid cells. Nature 517, 293–301 (2015).
McKenzie, A.N., Spits, H. & Eberl, G. Innate lymphoid cells in inflammation and immunity. Immunity 41, 366–374 (2014).
Spits, H. et al. Innate lymphoid cells--a proposal for uniform nomenclature. Nat. Rev. Immunol. 13, 145–149 (2013).
Bernink, J.H. et al. Human type 1 innate lymphoid cells accumulate in inflamed mucosal tissues. Nat. Immunol. 14, 221–229 (2013).
Daussy, C. et al. T-bet and Eomes instruct the development of two distinct natural killer cell lineages in the liver and in the bone marrow. J. Exp. Med. 211, 563–577 (2014).
Fuchs, A. et al. Intraepithelial type 1 innate lymphoid cells are a unique subset of IL-12- and IL-15-responsive IFN-γ-producing cells. Immunity 38, 769–781 (2013).
Klose, C.S. et al. Differentiation of type 1 ILCs from a common progenitor to all helper-like innate lymphoid cell lineages. Cell 157, 340–356 (2014).
Furusawa, J. et al. Critical role of p38 and GATA3 in natural helper cell function. J. Immunol. 191, 1818–1826 (2013).
Liang, H.E. et al. Divergent expression patterns of IL-4 and IL-13 define unique functions in allergic immunity. Nat. Immunol. 13, 58–66 (2012).
Hoyler, T., Connor, C.A., Kiss, E.A. & Diefenbach, A. T-bet and Gata3 in controlling type 1 and type 2 immunity mediated by innate lymphoid cells. Curr. Opin. Immunol. 25, 139–147 (2013).
Mjösberg, J. et al. The transcription factor GATA3 is essential for the function of human type 2 innate lymphoid cells. Immunity 37, 649–659 (2012).
Cella, M. et al. A human natural killer cell subset provides an innate source of IL-22 for mucosal immunity. Nature 457, 722–725 (2009).
Luci, C. et al. Influence of the transcription factor RORγt on the development of NKp46+ cell populations in gut and skin. Nat. Immunol. 10, 75–82 (2009).
Satoh-Takayama, N. et al. Microbial flora drives interleukin 22 production in intestinal NKp46+ cells that provide innate mucosal immune defense. Immunity 29, 958–970 (2008).
Sanos, S.L., Vonarbourg, C., Mortha, A. & Diefenbach, A. Control of epithelial cell function by interleukin-22-producing RORγt+ innate lymphoid cells. Immunology 132, 453–465 (2011).
Diefenbach, A., Colonna, M. & Koyasu, S. Development, differentiation, and diversity of innate lymphoid cells. Immunity 41, 354–365 (2014).
Cella, M., Otero, K. & Colonna, M. Expansion of human NK-22 cells with IL-7, IL-2, and IL-1β reveals intrinsic functional plasticity. Proc. Natl. Acad. Sci. USA 107, 10961–10966 (2010).
Hughes, T. et al. Stage 3 immature human natural killer cells found in secondary lymphoid tissue constitutively and selectively express the TH 17 cytokine interleukin-22. Blood 113, 4008–4010 (2009).
Hughes, T. et al. Interleukin-1β selectively expands and sustains interleukin-22+ immature human natural killer cells in secondary lymphoid tissue. Immunity 32, 803–814 (2010).
Klose, C.S.N. et al. A T-bet gradient controls the fate and function of CCR6−RORγt+ innate lymphoid cells. Nature 494, 261–265 (2013).
Vonarbourg, C. et al. Regulated expression of nuclear receptor RORγt confers distinct functional fates to NK cell receptor-expressing RORγt+ innate lymphocytes. Immunity 33, 736–751 (2010).
Sciumé, G. et al. Distinct requirements for T-bet in gut innate lymphoid cells. J. Exp. Med. 209, 2331–2338 (2012).
Bernink, J.H. et al. Interleukin-12 and -23 control plasticity of CD127+ group 1 and group 3 innate lymphoid cells in the intestinal lamina propria. Immunity 43, 146–160 (2015).
Crellin, N.K. et al. Regulation of cytokine secretion in human CD127+ LTi-like innate lymphoid cells by Toll-like receptor 2. Immunity 33, 752–764 (2010).
Sethi, S. & Murphy, T.F. Infection in the pathogenesis and course of chronic obstructive pulmonary disease. N. Engl. J. Med. 359, 2355–2365 (2008).
MacDonald, M., Korman, T., King, P., Hamza, K. & Bardin, P. Exacerbation phenotyping in chronic obstructive pulmonary disease. Respirology 18, 1280–1281 (2013).
Mallia, P. et al. Experimental rhinovirus infection as a human model of chronic obstructive pulmonary disease exacerbation. Am. J. Respir. Crit. Care Med. 183, 734–742 (2011).
Mohan, A. et al. Prevalence of viral infection detected by PCR and RT-PCR in patients with acute exacerbation of COPD: a systematic review. Respirology 15, 536–542 (2010).
Botelho, F.M. et al. IL-1α/IL-1R1 expression in chronic obstructive pulmonary disease and mechanistic relevance to smoke-induced neutrophilia in mice. PLoS One 6, e28457 (2011).
Kang, M.J. et al. IL-18 is induced and IL-18 receptor α plays a critical role in the pathogenesis of cigarette smoke-induced pulmonary emphysema and inflammation. J. Immunol. 178, 1948–1959 (2007).
Kearley, J. et al. Cigarette smoke silences innate lymphoid cell function and facilitates an exacerbated type I interleukin-33-dependent response to infection. Immunity 42, 566–579 (2015).
Iwasaki, A. & Pillai, P.S. Innate immunity to influenza virus infection. Nat. Rev. Immunol. 14, 315–328 (2014).
Lazarevic, V., Glimcher, L.H. & Lord, G.M. T-bet: a bridge between innate and adaptive immunity. Nat. Rev. Immunol. 13, 777–789 (2013).
Yagi, R. et al. The transcription factor GATA3 is critical for the development of all IL-7Rα-expressing innate lymphoid cells. Immunity 40, 378–388 (2014).
Monticelli, L.A. et al. Innate lymphoid cells promote lung-tissue homeostasis after infection with influenza virus. Nat. Immunol. 12, 1045–1054 (2011).
Regan, E.A. et al. Genetic epidemiology of COPD (COPDGene) study design. COPD 7, 32–43 (2010).
Ohne, Y. et al. IL-1 is a critical regulator of group 2 innate lymphoid cell function and plasticity. Nat. Immunol. http://dx.doi.org/10.1038/ni.3447 (25 April 2016).
Serafini, N. et al. Gata3 drives development of RORγt+ group 3 innate lymphoid cells. J. Exp. Med. 211, 199–208 (2014).
Zhong, C. et al. Group 3 innate lymphoid cells continuously require the transcription factor GATA-3 after commitment. Nat. Immunol. 17, 169–178 (2016).
Califano, D. et al. Transcription factor Bcl11b controls identity and function of mature type 2 innate lymphoid cells. Immunity 43, 354–368 (2015).
Bal, S.M. et al. Interleukin-1β, -4 and -12 control ILC2 fate in human airway inflammation. Nat. Immunol. http://dx.doi.org/10.1038/ni.3444 (25 April 2016).
Vignali, D.A.A. & Kuchroo, V.K. IL-12 family cytokines: immunological playmakers. Nat. Immunol. 13, 722–728 (2012).
Molofsky, A.B. et al. Interleukin-33 and Interferon-γ counter-regulate group 2 innate lymphoid cell activation during immune perturbation. Immunity 43, 161–174 (2015).
Duerr, C.U. et al. Type I interferon restricts type 2 immunopathology through the regulation of group 2 innate lymphoid cells. Nat. Immunol. 17, 65–75 (2016).
Moro, K. et al. Interferon and IL-27 antagonize the function of group 2 innate lymphoid cells and type 2 innate immune responses. Nat. Immunol. 17, 76–86 (2016).
Sojka, D.K. et al. Tissue-resident natural killer (NK) cells are cell lineages distinct from thymic and conventional splenic NK cells. eLife 3, e01659 (2014).
Bauer, C.M.T., Morissette, M.C. & Stämpfli, M.R. The influence of cigarette smoking on viral infections: translating bench science to impact COPD pathogenesis and acute exacerbations of COPD clinically. Chest 143, 196–206 (2013).
Byers, D.E. et al. Long-term IL-33-producing epithelial progenitor cells in chronic obstructive lung disease. J. Clin. Invest. 123, 3967–3982 (2013).
Neill, D.R. et al. Nuocytes represent a new innate effector leukocyte that mediates type-2 immunity. Nature 464, 1367–1370 (2010).
Moro, K. et al. Innate production of TH2 cytokines by adipose tissue-associated c-Kit+Sca-1+ lymphoid cells. Nature 463, 540–544 (2010).
Price, A.E. et al. Systemically dispersed innate IL-13-expressing cells in type 2 immunity. Proc. Natl. Acad. Sci. USA 107, 11489–11494 (2010).
Roberts, M.E.P. et al. CD4+ T-Cell profiles and peripheral blood ex-vivo responses to T-cell directed stimulation delineate COPD phenotypes. J. COPD Found. 2, 268–280 (2016).
Acknowledgements
We thank the MedImmune Flow Cytometry core for all cell sorting; the MedImmune histology core for embedding tissues; the LAR staff for maintaining the experimental mice; M. Stämpfli for expertise and guidance in establishing a smoking system at MedImmune; A. Gonzales for running and maintaining the system for in-house exposure to cigarette smoke; the C. Lopez laboratory (University of Pennsylvania, School of Veterinary Medicine) for influenza virus strain PR8; M. Snaith for help with generating the ST2-GFP reporter mouse; M.E.P. Roberts for facilitating the collaboration with National Jewish Health; C. Schnell, T. Thorn and the rest of the team at NJH for commitment and contributions to this collaborative effort; and J. Jönsson and K. Jansner for histological work and image processing and analysis.
Author information
Authors and Affiliations
Contributions
J.S.S., J.K. and A.A.H. planned all experiments; J.S.S. and J.K. executed and analyzed all experiments; A.M.C., L.Y., G.H.P. and A.A.B. planned and executed specific experiments; C.S., M.M. and J.S.E. cut, stained and analyzed all histology sections and developed the algorithms for analysis of ILC location; L.Y. analyzed and generated the statistical data and graphs for the COPDGene study; G.H.P. and C.A.H. provided Tbx21−/− mice and reagents; R.B. provided blood samples from patients with COPD and control subjects; C.A.H., J.S.E., R.K. and A.A.H. provided feedback and edits; and J.S.S. and A.A.H. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
J.S.S., J.K., A.M.C., L.Y., A.A.B., R.K. and A.A.H. are employed by and shareholders of MedImmune, and C.S., M.M., G.H.P., C.A.H., R.B. and J.S.E. have received funding from MedImmune.
Integrated supplementary information
Supplementary Figure 1 Generation of ST2-GFP reporter mice and characterization of ILC1 cells following infection with influenza virus.
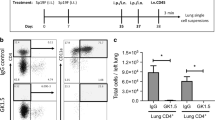
(a) ST2-GFP reporter mice were generated as described in Methods. (b) Representative flow cytometric plots demonstrating Lineage vs. ST2-GFP staining in lung of littermate control or ST2-GFP reporter mice; and expression of CD90, IL-7Rα, CD44 and CD25 on Lin- ST2-GFP+ cells. Representative flow cytometric plots comparing ILC populations in naive lung, infected lung and draining lymph nodes at Day 7 post-infection (c). Representative flow cytometry plots comparing ILC populations in the bronchoalveolar lavage fluid from naive and infected mice (d). Live/dead fixable blue (e) or Annexin V (f) staining on ILCs in naïve and infected lungs at Day 7 post-infection. (g) Expression of GATA-3, T-bet, CD44, CD25, IL-7rα, cKit and ICOS on lung-resident ST2+ (red) or IL-18rα+ (blue) ILCs at Day 7 post-infection. (h) Percentage of lung ILCs positive for Ki67. (i) IL-5, IL-13 and IFN-γ levels in culture supernatants from lung ILCs isolated from the lung at day 7 post-infection and stimulated as indicated for 24 hours. Data are expressed as Mean±S.E.M., b-h are representative of 2 independent experiments with 3-5 mice per group; 1l is representative of 2 separate experiments with 9 mice/group. *P<.01 according to the Mann-whitney Wilcoxon test.
Supplementary Figure 2 Bacterial infection and/or exposure cigarette smoke trigger(s) phenotypic changes in lung-resident ILC2 cells.
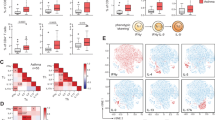
(a-c) Quantification of MFI for IL-18Rα, Tbet and IL-12Rβ2 on lung-resident ILCs in naïve mice and mice infected with Staphylococcus aureus (S.aureus) at day 5 post-challenge. (d) Representative flow cytometric plots of GATA-3 expression in naive mice or mice infected with nontypeable Haemophilus influenzae (NTHi). at day 2 post-infection. (e-f) Representative flow cytometric plots and MFI of IL-12Rβ2 expression on lung-resident ILCs in naive mice and mice infected with NTHi at day 5 post-infection (g). Percentage of cells expressing GATA-3 in naïve mice, mice infected with influenza, or mice exposed to cigarette smoke prior to influenza infection. (h) Representative flow cytometric plots of ST2 and IL-18Rα expression on lung-resident ILCs in naive mice, influenza-infected mice or mice exposed to cigarette smoke prior to influenza infection. Correlation (i) and percentage (j) of ILCs expressing ST2 or IL-18Rα in naive mice, influenza-infected mice or mice exposed to cigarette smoke prior to influenza infection. (k-l) GATA-3 and T-bet expression in lung-resident ILCs from naive mice, influenza-infected mice or mice exposed to cigarette smoke prior to influenza infection. *p<.01, **p<.001. Data in a-f are representative of 2-3 independent experiments with ≥5 mice per group. Data in g are pooled from three independent experiments with 3-15 mice/group. Data in h-l are representative of 3-4 independent experiments with ≥5 mice per group. *p<.01, **p<.001.
Supplementary Figure 3 IL-12 and IL-18 induce IFN-γ production by ILC2 cells in vitro and in vivo.
(a) Representative flow cytometric plots of ILCs following double depletion of lineage positive cells. ILCs were defined as CD45+, viable, CD3-, CD49b-, Lin- CD44+ CD90+ IL-7rα+. Levels of IL-5 (b) and IL-13 (c) as measured in the culture supernatants after 4 days of culture with IL-12 and IL-18. MFI of GATA-3 (d), ST2 (e), T-bet (f) and IL-18Rα (g) on lung-resident ILCs from mice treated intranasally as indicated. (h-j) Representative flow cytometric plots showing IL-5 and IFN-g on ILCs were purified from lungs of mice treated as indicated. Data in a-c are representative of three independent experiments, where ILC2 cells were pooled from n≥8 mice. Data in d-g are representative of two independent experiments. For h-j, ILCs were pooled from n≥5 mice per treatment group. *p<.01, **p<.001.
Supplementary Figure 4 GFP+ ILC2 cells adopt an ILC1 phenotype following infection with influenza virus.
(a) ST2+IL-18Rα- ILC2 were sorted and transferred in RAG/γc-/- mice, which were subsequently infected with influenza A. Representative flow cytometric plots showing GATA-3 (b), IL-18Rα (c) and IL-12Rβ2 (d) expression on adoptively transferred ILC2 cells 10 days after virus infection. Representative flow cytometric plots showing IFNγ (d) or IL-13 (e) expression on adoptively transferred ILC2 cells purified from the lung and stimulated overnight with IL-12 and IL-18 (d) or IL-33 (e). MFI of GATA-3 (f), IL-18Rα (g) and IL-12Rβ2 (h) on adoptively transferred ILCs. (i) IFN-γ levels in the supernatants of ILCs cultured overnight in indicated conditions. Data are expressed as Mean and are representative of 2 independent experiments with 5-9 mice per group (g-i) or from 2 independent experiments where each group was pooled (j). ***p<0.0001. All data was analyzed at day 10 post-infection.
Supplementary Figure 5 Rate of influenza-virus infectivity and imaging of transferred GFP+ ILC2 cells in tissue.
(a) Quantification of influenza A staining in RAG/γc-/- deficient mice at day 10 post-infection. Data are expressed as Mean and are representative of 2 independent experiments with 8-11 mice/group, ***p<0.0001. (b) Positive control of GFP immunostaining with brown DAB chromogen in an infected RAG/γc-/- mouse that received GFP+ ILC2 cells. (c) Negative control with the same staining protocol in a virus-infected mouse that had not received any GFP cells. (d) Double in situ hybridization (ISH) with simultaneous visualization of IL-12 and IL18 mRNA by green and red chromogen, respectively. The image is from 6 d post infection and influenza virus particles are immunostained with brown DAB chromogen. (e) Positive ISH control with probe for the eukaryote house-keeping gene PPIB. (f) Negative control with the non-eukaryote bacterial gene DapB. (g,h) Positive staining control for IL-12 and IL-18 alone, respectively. Scale bars: b-h=20μm.
Supplementary Figure 6 T-bet promotes IFN-γ production by ILC2 cells and effects of CD90 depletion on lung-resident cells.
Representative flow cytometric plots (a) and quantification (b) of GATA-3 expression in lung-resident ILCs in C57BL/6 and Tbx21-/- mice after intranasal treatment with PBS or IL-12 and IL-18. Representative flow cytometric plots (c) of ST2 and IL-18Rα expression, quantified in (d), in lung-resident ILCs in C57BL/6 and Tbx21-/- mice treated intranasally with PBS or IL-12 and IL-18. Representative flow cytometric plots (e) and quantification (f) of IL-12Rβ2 expression in lung-resident ILCs in C57BL/6 and Tbx21-/- mice treated intranasally with PBS or IL-12 and IL-18. (g) Intracellular IFN-γ expression in purified ILCs from PBS- or IL-12/IL-18-treated C57BL/6 and Tbx21-/- mice that were stimulated overnight with IL-12 and IL-18. (h) IFN-γ levels in culture supernatants from purified ILCs stimulated overnight as indicated. Representative flow cytometric plots (i) showing Lin- CD90+ cells (gated on CD45+, viable, CD3- CD49b- cells) in mice treated with isotype control or anti-CD90.2 antibody. (j) Quantification of the percentage of ILCs in naive and infected mice treated with isotype control or anti-CD90.2 antibody. Representative flow cytometric plots (k) showing NK cell populations (defined as viable, CD45+, CD3- CD49b+ cells) in naive and infected SCID mice treated with isotype control or anti CD90.2 antibody. (l) Quantification of NK cells in the lungs of naïve and infected SCID mice treated with isotype control or anti CD90.2 antibody. Representative flow cytometric plots (m) showing mesenchymal stem cell populations (mSC, defined as viable, CD45-, CD90+ cells) in naive and infected mice treated with isotype control or anti CD90.2 antibody. (n) Quantification of mSC cells in the lungs of naïve and infected mice treated with isotype control or anti CD90.2 antibody. (o-p) CD166 and Sca-1 expression on CD90+ CD45- cells in the lung. (q) ILC2 and ILC1 cells were purified from mice treated with IL-33 or IL-33 +IL-12+IL-18, respectively, and transferred into RAG/γc deficient mice. Data are shown as Mean and are representative of 2 independent experiments with 4-9 mice/group (a-g) or 7-10 mice/group (j-n) or groups were pooled from 2 independent experiments (h). *p<.01, **p<.001, ***p<.0001.
Supplementary Figure 7 Flow cytometry gating strategy for ILC2 cells from healthy human donors.
Human PBMCs were pooled from 3-4 healthy donors and enriched for NK cells/ILCs using an NK cell enrichment kit. Purity of ILCs in post-NK-enrichment demonstrated by representative flow cytometric plots showing pre-sort (a) and post-sort (b) samples for in vitro culture of human ILC2 cells, defined here as CD45+ Viable CD3- CD19- Lin- IL-7Rα+ CRTH2+ CD161+. (c) Representative flow cytometric plots demonstrating post-culture purity human ILC2 cells cultured with IL-2 and IL-33.
Supplementary Figure 8 Flow cytometry gating strategy for the identification and quantification of ILC subsets in patients with COPD.
(a) Representative flow cytometric plots demonstrating ILC gating strategy for identifying ILCs in the peripheral blood of non-smokers (aged matched healthy controls), healthy smokers or COPD patients. Samples were depleted of T and B cells and ILCs were defined as CD45+ Viable CD3- CD19- Lin- IL-7Rα+ CD56-. (b) Correlative analysis between FEV1/FVC and % ILC1 cells. (c) Correlative analysis between FEV1 predicted and % ILC2 cells. (d) Correlative analysis between FEV1/FVC and % ILC2 cells in the peripheral blood of non-smokers, healthy smokers and COPD patients. (e) Quantification of the sum of % ILC1 and ILC2 cells in non-smokers, healthy smokers and COPD patients, dotted line indicates the mean ± SD across all groups; 42.3%±13.1%. Group sizes: b-d GOLD I=4; GOLD II=11; GOLD III=12; GOLD IV=14; e Non-smokers=11; Smokers=18; total COPD=42.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–8 and Supplementary Tables 1 and 2 (PDF 1371 kb)
Rights and permissions
About this article
Cite this article
Silver, J., Kearley, J., Copenhaver, A. et al. Inflammatory triggers associated with exacerbations of COPD orchestrate plasticity of group 2 innate lymphoid cells in the lungs. Nat Immunol 17, 626–635 (2016). https://doi.org/10.1038/ni.3443
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ni.3443
This article is cited by
-
A diversity of novel type-2 innate lymphoid cell subpopulations revealed during tumour expansion
Communications Biology (2024)
-
Advances in lung ischemia/reperfusion injury: unraveling the role of innate immunity
Inflammation Research (2024)
-
Physiological and immunological barriers in the lung
Seminars in Immunopathology (2024)
-
Butyrate inhibits iILC2-mediated lung inflammation via lung-gut axis in chronic obstructive pulmonary disease (COPD)
BMC Pulmonary Medicine (2023)
-
Development and validation of an in-hospital mortality risk prediction model for patients with severe community-acquired pneumonia in the intensive care unit
BMC Pulmonary Medicine (2023)