Abstract
The NLRP3 inflammasome responds to microbes and danger signals by processing and activating proinflammatory cytokines, including interleukin 1β (IL-1β) and IL-18. We found here that activation of the NLRP3 inflammasome was restricted to interphase of the cell cycle by NEK7, a serine-threonine kinase previously linked to mitosis. Activation of the NLRP3 inflammasome required NEK7, which bound to the leucine-rich repeat domain of NLRP3 in a kinase-independent manner downstream of the induction of mitochondrial reactive oxygen species (ROS). This interaction was necessary for the formation of a complex containing NLRP3 and the adaptor ASC, oligomerization of ASC and activation of caspase-1. NEK7 promoted the NLRP3-dependent cellular inflammatory response to intraperitoneal challenge with monosodium urate and the development of experimental autoimmune encephalitis in mice. Our findings suggest that NEK7 serves as a cellular switch that enforces mutual exclusivity of the inflammasome response and cell division.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Gross, O., Thomas, C.J., Guarda, G. & Tschopp, J. The inflammasome: an integrated view. Immunol. Rev. 243, 136–151 (2011).
Zhou, R., Yazdi, A.S., Menu, P. & Tschopp, J. A role for mitochondria in NLRP3 inflammasome activation. Nature 469, 221–225 (2011).
Subramanian, N., Natarajan, K., Clatworthy, M.R., Wang, Z. & Germain, R.N. The adaptor MAVS promotes NLRP3 mitochondrial localization and inflammasome activation. Cell 153, 348–361 (2013).
Misawa, T. et al. Microtubule-driven spatial arrangement of mitochondria promotes activation of the NLRP3 inflammasome. Nat. Immunol. 14, 454–460 (2013).
Roig, J., Mikhailov, A., Belham, C. & Avruch, J. Nercc1, a mammalian NIMA-family kinase, binds the Ran GTPase and regulates mitotic progression. Genes Dev. 16, 1640–1658 (2002).
Belham, C. et al. A mitotic cascade of NIMA family kinases. Nercc1/Nek9 activates the Nek6 and Nek7 kinases. J. Biol. Chem. 278, 34897–34909 (2003).
Yissachar, N., Salem, H., Tennenbaum, T. & Motro, B. Nek7 kinase is enriched at the centrosome, and is required for proper spindle assembly and mitotic progression. FEBS Lett. 580, 6489–6495 (2006).
O'Regan, L. & Fry, A.M. The Nek6 and Nek7 protein kinases are required for robust mitotic spindle formation and cytokinesis. Mol. Cell. Biol. 29, 3975–3990 (2009).
Bertran, M.T. et al. Nek9 is a Plk1-activated kinase that controls early centrosome separation through Nek6/7 and Eg5. EMBO J. 30, 2634–2647 (2011).
Kim, S., Lee, K. & Rhee, K. NEK7 is a centrosomal kinase critical for microtubule nucleation. Biochem. Biophys. Res. Commun. 360, 56–62 (2007).
Salem, H. et al. Nek7 kinase targeting leads to early mortality, cytokinesis disturbance and polyploidy. Oncogene 29, 4046–4057 (2010).
Kim, S., Kim, S. & Rhee, K. NEK7 is essential for centriole duplication and centrosomal accumulation of pericentriolar material proteins in interphase cells. J. Cell Sci. 124, 3760–3770 (2011).
Wang, T. et al. Real-time resolution of point mutations that cause phenovariance in mice. Proc. Natl. Acad. Sci. USA 112, E440–E449 (2015).
Kayagaki, N. et al. Non-canonical inflammasome activation targets caspase-11. Nature 479, 117–121 (2011).
Jacobs, C.A. et al. Experimental autoimmune encephalomyelitis is exacerbated by IL-1α and suppressed by soluble IL-1 receptor. J. Immunol. 146, 2983–2989 (1991).
Martin, D. & Near, S.L. Protective effect of the interleukin-1 receptor antagonist (IL-1ra) on experimental allergic encephalomyelitis in rats. J. Neuroimmunol. 61, 241–245 (1995).
Muñoz-Planillo, R. et al. K+ efflux is the common trigger of NLRP3 inflammasome activation by bacterial toxins and particulate matter. Immunity 38, 1142–1153 (2013).
Lee, G.S. et al. The calcium-sensing receptor regulates the NLRP3 inflammasome through Ca2+ and cAMP. Nature 492, 123–127 (2012).
Fernandes-Alnemri, T. et al. The pyroptosome: a supramolecular assembly of ASC dimers mediating inflammatory cell death via caspase-1 activation. Cell Death Differ. 14, 1590–1604 (2007).
Broz, P., von Moltke, J., Jones, J.W., Vance, R.E. & Monack, D.M. Differential requirement for caspase-1 autoproteolysis in pathogen-induced cell death and cytokine processing. Cell Host Microbe 8, 471–483 (2010).
Lu, A. et al. Unified polymerization mechanism for the assembly of ASC-dependent inflammasomes. Cell 156, 1193–1206 (2014).
Compan, V. et al. Cell volume regulation modulates NLRP3 inflammasome activation. Immunity 37, 487–500 (2012).
Pelegrin, P., Barroso-Gutierrez, C. & Surprenant, A. P2X7 receptor differentially couples to distinct release pathways for IL-1β in mouse macrophage. J. Immunol. 180, 7147–7157 (2008).
Matsubayashi, T., Sugiura, H., Arai, T., Oh-Ishi, T. & Inamo, Y. Anakinra therapy for CINCA syndrome with a novel mutation in exon 4 of the CIAS1 gene. Acta Paediatr. 95, 246–249 (2006).
Aksentijevich, I., Remmers, E.F., Goldbach-Mansky, R., Reiff, A. & Kastner, D.L. Mutational analysis in neonatal-onset multisystem inflammatory disease: comment on the articles by Frenkel et al. and Saito et al. Arthritis Rheum. 54, 2703–2704 (2006).
Jesus, A.A. et al. Phenotype-genotype analysis of cryopyrin-associated periodic syndromes (CAPS): description of a rare non-exon 3 and a novel CIAS1 missense mutation. J. Clin. Immunol. 28, 134–138 (2008).
Kandli, M., Feige, E., Chen, A., Kilfin, G. & Motro, B. Isolation and characterization of two evolutionarily conserved murine kinases (Nek6 and Nek7) related to the fungal mitotic regulator, NIMA. Genomics 68, 187–196 (2000).
Vassilev, L.T. et al. Selective small-molecule inhibitor reveals critical mitotic functions of human CDK1. Proc. Natl. Acad. Sci. USA 103, 10660–10665 (2006).
Weinberg, S.E., Sena, L.A. & Chandel, N.S. Mitochondria in the regulation of innate and adaptive immunity. Immunity 42, 406–417 (2015).
Cooper, G.M. in The Cell: A Molecular Approach (Sinauer Associates, Sunderland, Massachusetts, 2000).
Eder, C. Mechanisms of interleukin-1β release. Immunobiology 214, 543–553 (2009).
Li, Q. & Engelhardt, J.F. Interleukin-1beta induction of NFκB is partially regulated by H2O2-mediated activation of NFκB-inducing kinase. J. Biol. Chem. 281, 1495–1505 (2006).
Bertram, C. & Hass, R. Cellular responses to reactive oxygen species-induced DNA damage and aging. Biol. Chem. 389, 211–220 (2008).
Orthwein, A. et al. Mitosis inhibits DNA double-strand break repair to guard against telomere fusions. Science 344, 189–193 (2014).
Georgel, P., Du, X., Hoebe, K. & Beutler, B. ENU mutagenesis in mice. Methods Mol. Biol. 415, 1–16 (2008).
Lyons, J.A., Ramsbottom, M.J. & Cross, A.H. Critical role of antigen-specific antibody in experimental autoimmune encephalomyelitis induced by recombinant myelin oligodendrocyte glycoprotein. Eur. J. Immunol. 32, 1905–1913 (2002).
Acknowledgements
We thank H. Zaki (University of Texas Southwestern Medical Center) for Nlrp3−/− (B6.129S6-Nlrp3tm1Bhk/J) mice; and F. Shao (National Institute of Biological Sciences, Beijing, China) for the NLRC4 plasmid. Supported by the US National Institutes of Health (U19 AI100627).
Author information
Authors and Affiliations
Contributions
H.S. and B.B. designed the study and analyzed data, with suggestions from N.L.M. and R.J.U.; H.S., Y.W., L.Su, D.C., V.V.K. and J.C.M. performed experiments; H.S. and Y.W. identified the Cuties phenotype; X.L., X.Z., M.T. and M.F. generated the Nek7-knockout mice; D.P., C.H.B., S.H., S.L., L.Sc, J.Q. and Q.S. performed genome mapping and genotyping; H.S., J.R. and S.A. maintained the Cuties mice; H.S., P.J., E.M.Y.M. and B.B. edited the figures; and H.S., E.M.Y.M. and B.B. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
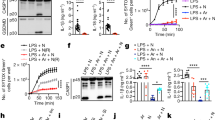
Supplementary Figure 1 Specific, functional defect in activation of the NLRP3 inflammasome in Cuties mice.
ELISA analysis of IL-1β secretion by (a) BMDM and (b) BMDC primed with LPS and treated with the indicated stimuli (n = 3 mice per genotype). (c,d) Time course of IL-1β secretion, IL-18 secretion, and pyroptosis of peritoneal macrophages primed with LPS and treated with (c) nigericin or (d) ATP (n = 4 mice per genotype). P values: Nek7+/+ vs. Nek7+/Cu (gray); Nek7+/+ vs. Nek7Cu/Cu (magenta); Nek7+/+ vs. Nlrp3-/- (teal); Nek7Cu/Cu vs. Nlrp3-/- (blue). * P≤0.05; ** P≤0.01; *** P≤0.001; **** P≤0.0001 (unpaired, two-tailed Student’s t test). (e) TNF and (f) IL-6 secretion by peritoneal macrophages treated with the indicated stimuli (n = 4 mice per genotype). Results are representative of two independent experiments (mean and s.d. shown).
Supplementary Figure 2 Cuties mice have normal blood cell populations and macrophage development, peritoneal recruitment and apoptosis.
(a) Flow cytometric analysis of the frequencies of myeloid cells and lymphocytes in peripheral blood of Cuties mice. Data points represent individual mice. (b) Bone marrow progenitor colony forming unit assay. Colonies formed from bone marrow cells cultured in methylcellulose base media with M-CSF (upper panel) and quantitated in the lower panel (n = 4 mice per genotype). (c) Flow cytometric analysis of the number of peritoneal exudate cells (PECs), neutrophils (Ly6G+ F4/80-), and monocytes/macrophages (F4/80+) in the peritoneum of Nek7+/+ (filled circles) or Nek7Cu/Cu mice (open circles) 6 h after intraperitoneal injection of IL-1β. Data points represent individual mice. (d) Flow cytometric analysis of untreated or LPS-primed macrophages stained with Annexin V and the viability dye 7-AAD. Results are representative of two independent experiments (mean and s.d. shown in a and b; horizontal line represents mean in c).
Supplementary Figure 3 Confirmation of the Cuties phenotype in mouse macrophages and human monocytes.
(a,b,d) Endogenous NEK7 expression was knocked down using siRNAs. (a) ELISA analysis of IL-1β secretion by J774A.1 cells primed with LPS and treated with nigericin or ATP. (b) HEK293T cells were transfected with NLRP3 inflammasome components and treated with nigericin or ATP. Immunoblots showing expression of the indicated proteins in cell lysates (Lys) and culture supernatants (Sup). IL-1β maturation was assessed by p17-Flag immunoblot of the supernatant. (c) Nek7-/- mice were generated by CRISPR/Cas9 targeting and peritoneal macrophages were assayed by ELISA for IL-1β secretion after priming with LPS and treatment with the indicated inflammasome stimuli (n = 3 mice per genotype). (Inset) Immunoblot showing NEK7 expression in peritoneal macrophages. (d) ELISA analysis of IL-1β secretion by THP1 cells primed with LPS and treated with nigericin or ATP. (e,f) Endogenous NEK7 expression was knocked down using shRNAs in primary human monocytes from healthy donor peripheral blood. ELISA analysis of (e) IL-1β and (f) IL-6 secretion in response to LPS priming and nigericin treatment. (e, top) Immunoblot showing NEK7 expression in primary human monocytes after shRNA knockdown. ** P≤0.01; *** P≤0.001; **** P≤0.0001 (unpaired, two-tailed Student’s t test). Results are representative of two independent experiments in which treatments were performed in triplicate (a,d), two independent experiments (b,c) and three independent experiments in which treatments were performed in triplicate (e,f); data in a and c-f presented as mean and s.d.. N.C, non-targeting control siRNA or shRNA.
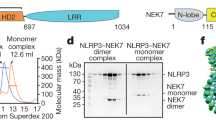
Supplementary Figure 4 Direct interaction between NEK7 and NLRP3 and in vitro NEK7 kinase assay.
(a,b) The indicated recombinant proteins were incubated together and amylose magnetic beads were used to precipitate MBP or MBP tagged NEK7 proteins (MBP PD). Complexes were analyzed by immunoblotting with the indicated antibodies. CBB, Coomassie Brilliant Blue. (c) Recombinant MBP-NEK7 or MBP-NEK7(K64M) was incubated with β-casein and ATP. NEK7 kinase activity was measured using an ADP-based phosphatase coupled kinase assay and plotted relative to the activity measured without any MBP or MBP conjugated protein (Mock). * P≤0.05 (unpaired, two-tailed Student’s t test). Results are representative of two independent experiments (a and b) or two independent experiments in which assays were performed in triplicate (c; mean and s.d. shown).
Supplementary Figure 5 NEK7 phosphorylation enhances its binding to NLRP3 and promotes inflammasome activation.
(a) Endogenous NEK7-NLRP3 association in LPS-primed J774A.1 cells treated with the indicated inhibitors, stimulated with nigericin, and analyzed by immunoprecipitation and immunoblot. (b,c) Endogenous NEK7-NLRP3 association was analyzed by immunoprecipitation and immunoblot. The phosphorylation state of NEK7 was analyzed using Phos-tag SDS-PAGE. (b) J774A.1 cells primed with LPS and stimulated with ATP were lysed and treated with calf intestinal alkaline phosphatase (CIP). (c) J774A.1 cells primed with LPS and stimulated with ATP together with N-acetylcysteine (NAC). (d) ELISA analysis of IL-1β in the culture supernatants of J774A.1 cells treated as in c. The means of triplicate samples are plotted. Results are representative of two independent experiments.
Supplementary Figure 6 NEK6 is not required for activation of the NLRP3 inflammasome.
(a) Wild type peritoneal macrophages were primed with LPS and stimulated with nigericin or ATP. The interaction between endogenous NEK6 or NEK7 and NLRP3 was analyzed by immunoprecipitation and immunoblot. (b) Immunoblot showing NEK6 expression in lysates of peritoneal macrophages from mice with a Nek6 mutation, a T to C transition at 38569737 bp on Chr 2 affecting the invariant GU dinucleotide of the intron 7 donor splice site (+2 bp from the exon 7 boundary). (c) ELISA analysis of IL-1β secretion by peritoneal macrophages primed with LPS and treated with the indicated inflammasome stimuli (n = 3 mice per genotype). Results are representative of two independent experiments (mean and s.d. shown in c).
Supplementary Figure 7 The NLRP3 inflammasome is not activated by lysates of mitotic cells.
(a) NEK7-NLRP3 interaction during mitosis. HEK293T cells were transfected as indicated (above lanes in right panel) and then treated with RO-3306 for 20 h before release in fresh media for the indicated times. Flow cytometric analysis of phosphorylated histone H3-positive mitotic cells (left); the percentage of mitotic cells is indicated. The interaction between NEK7 and NLRP3 was analyzed by immunoprecipitation and immunoblot (right). (b) ELISA analysis of IL-1b secretion by peritoneal macrophages primed with LPS and treated with lysates of non-synchronized cycling HEK293T cells (cytosol extract), or lysates of mitotic HEK293T cells (mitotic cell extract) (mean and s.d. of n = 4 mice per genotype). Results are representative of two independent experiments.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–7 (PDF 1153 kb)
Rights and permissions
About this article
Cite this article
Shi, H., Wang, Y., Li, X. et al. NLRP3 activation and mitosis are mutually exclusive events coordinated by NEK7, a new inflammasome component. Nat Immunol 17, 250–258 (2016). https://doi.org/10.1038/ni.3333
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ni.3333
This article is cited by
-
The neuropeptide CGRP enters the macrophage cytosol to suppress the NLRP3 inflammasome during pulmonary infection
Cellular & Molecular Immunology (2023)
-
Cryo-EM structures of the active NLRP3 inflammasome disc
Nature (2023)
-
Theaflavin mitigates acute gouty peritonitis and septic organ injury in mice by suppressing NLRP3 inflammasome assembly
Acta Pharmacologica Sinica (2023)
-
Mitochondrial damage activates the NLRP10 inflammasome
Nature Immunology (2023)
-
Chicken cathelicidin-2 promotes NLRP3 inflammasome activation in macrophages
Veterinary Research (2022)