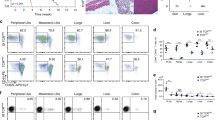
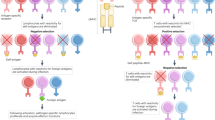
Abstract
Studies of repertoires of mouse monoclonal CD4+ T cells have revealed several mechanisms of self-tolerance; however, which mechanisms operate in normal repertoires is unclear. Here we studied polyclonal CD4+ T cells specific for green fluorescent protein expressed in various organs, which allowed us to determine the effects of specific expression patterns on the same epitope-specific T cells. Peptides presented uniformly by thymic antigen-presenting cells were tolerated by clonal deletion, whereas peptides excluded from the thymus were ignored. Peptides with limited thymic expression induced partial clonal deletion and impaired effector T cell potential but enhanced regulatory T cell potential. These mechanisms were also active for T cell populations specific for endogenously expressed self antigens. Thus, the immunotolerance of polyclonal CD4+ T cells was maintained by distinct mechanisms, according to self-peptide expression patterns.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
References
Xing, Y. & Hogquist, K.A. T-cell tolerance: central and peripheral. Cold Spring Harb. Perspect. Biol. 4, a006957 (2012).
Wing, K. & Sakaguchi, S. Regulatory T cells exert checks and balances on self tolerance and autoimmunity. Nat. Immunol. 11, 7–13 (2010).
Semana, G., Gausling, R., Jackson, R.A. & Hafler, D.A. T cell autoreactivity to proinsulin epitopes in diabetic patients and healthy subjects. J. Autoimmun. 12, 259–267 (1999).
Moon, J.J. et al. Quantitative impact of thymic selection on Foxp3+ and Foxp3− subsets of self-peptide/MHC class II-specific CD4+ T cells. Proc. Natl. Acad. Sci. USA 108, 14602–14607 (2011).
Yu, W. et al. Clonal deletion prunes but does not eliminate self-specific αβ CD8+ T lymphocytes. Immunity 42, 929–941 (2015).
Maeda, Y. et al. Detection of self-reactive CD8+ T cells with an anergic phenotype in healthy individuals. Science 346, 1536–1540 (2014).
Ohashi, P.S. et al. Ablation of “tolerance” and induction of diabetes by virus infection in viral antigen transgenic mice. Cell 65, 305–317 (1991).
Oldstone, M.B., Nerenberg, M., Southern, P., Price, J. & Lewicki, H. Virus infection triggers insulin-dependent diabetes mellitus in a transgenic model: role of anti-self (virus) immune response. Cell 65, 319–331 (1991).
Jenkins, M.K. & Schwartz, R.H. Antigen presentation by chemically modified splenocytes induces antigen-specific T cell unresponsiveness in vitro and in vivo. J. Exp. Med. 165, 302–319 (1987).
Akkaraju, S. et al. A range of CD4 T cell tolerance: partial inactivation to organ-specific antigen allows nondestructive thyroiditis or insulitis. Immunity 7, 255–271 (1997).
Zehn, D. & Bevan, M.J. T cells with low avidity for a tissue-restricted antigen routinely evade central and peripheral tolerance and cause autoimmunity. Immunity 25, 261–270 (2006).
Hataye, J., Moon, J.J., Khoruts, A., Reilly, C. & Jenkins, M.K. Naive and memory CD4+ T cell survival controlled by clonal abundance. Science 312, 114–116 (2006).
Marzo, A.L. et al. Initial T cell frequency dictates memory CD8+ T cell lineage commitment. Nat. Immunol. 6, 793–799 (2005).
Bautista, J.L. et al. Intraclonal competition limits the fate determination of regulatory T cells in the thymus. Nat. Immunol. 10, 610–617 (2009).
Bouneaud, C., Kourilsky, P. & Bousso, P. Impact of negative selection on the T cell repertoire reactive to a self-peptide: a large fraction of T cell clones escapes clonal deletion. Immunity 13, 829–840 (2000).
Nelson, R.W. et al. T cell receptor cross-reactivity between similar foreign and self peptides influences naive cell population size and autoimmunity. Immunity 42, 95–107 (2015).
Reich, M. et al. GenePattern 2.0. Nat. Genet. 38, 500–501 (2006).
Hara, M. et al. Transgenic mice with green fluorescent protein-labeled pancreatic beta-cells. Am. J. Physiol. Endocrinol. Metab. 284, E177–E183 (2003).
Anderson, M.S. et al. Projection of an immunological self shadow within the thymus by the aire protein. Science 298, 1395–1401 (2002).
Carreres, M.I. et al. Transcription factor Foxd1 is required for the specification of the temporal retina in mammals. J. Neurosci. 31, 5673–5681 (2011).
Kobayashi, A. et al. Identification of a multipotent self-renewing stromal progenitor population during mammalian kidney organogenesis. Stem Cell Rep. 3, 650–662 (2014).
Ben-Yehudah, A. et al. Specific dynamic and noninvasive labeling of pancreatic beta cells in reporter mice. Genesis 43, 166–174 (2005).
Kaplan, D.H. et al. Autocrine/paracrine TGFβ1 is required for the development of epidermal Langerhans cells. J. Exp. Med. 204, 2545–2552 (2007).
Heng, T.S., Painter, M.W. & Immunological Genome Project Consortium. The Immunological Genome Project: networks of gene expression in immune cells. Nat. Immunol. 9, 1091–1094 (2008).
Gong, S. et al. A gene expression atlas of the central nervous system based on bacterial artificial chromosomes. Nature 425, 917–925 (2003).
Crawford, F., Kozono, H., White, J., Marrack, P. & Kappler, J. Detection of antigen-specific T cells with multivalent soluble class II MHC covalent peptide complexes. Immunity 8, 675–682 (1998).
Warren, H.S., Vogel, F.R. & Chedid, L.A. Current status of immunological adjuvants. Annu. Rev. Immunol. 4, 369–388 (1986).
Portnoy, D.A., Auerbuch, V. & Glomski, I.J. The cell biology of Listeria monocytogenes infection: the intersection of bacterial pathogenesis and cell-mediated immunity. J. Cell Biol. 158, 409–414 (2002).
Cordier, A.C. & Haumont, S.M. Development of thymus, parathyroids, and ultimo-branchial bodies in NMRI and nude mice. Am. J. Anat. 157, 227–263 (1980).
Schaefer, B.C., Schaefer, M.L., Kappler, J.W., Marrack, P. & Kedl, R.M. Observation of antigen-dependent CD8+ T-cell/ dendritic cell interactions in vivo. Cell. Immunol. 214, 110–122 (2001).
Okabe, M., Ikawa, M., Kominami, K., Nakanishi, T. & Nishimune, Y. 'Green mice' as a source of ubiquitous green cells. FEBS Lett. 407, 313–319 (1997).
Lindquist, R.L. et al. Visualizing dendritic cell networks in vivo. Nat. Immunol. 5, 1243–1250 (2004).
Panneck, A.R. et al. Cholinergic epithelial cell with chemosensory traits in murine thymic medulla. Cell Tissue Res. 358, 737–748 (2014).
Tallini, Y.N. et al. BAC transgenic mice express enhanced green fluorescent protein in central and peripheral cholinergic neurons. Physiol. Genomics 27, 391–397 (2006).
Snippert, H.J. et al. Intestinal crypt homeostasis results from neutral competition between symmetrically dividing Lgr5 stem cells. Cell 143, 134–144 (2010).
Gardner, J.M. et al. Deletional tolerance mediated by extrathymic Aire-expressing cells. Science 321, 843–847 (2008).
Pepper, M., Pagan, A.J., Igyarto, B.Z., Taylor, J.J. & Jenkins, M.K. Opposing signals from the Bcl6 transcription factor and the interleukin-2 receptor generate T helper 1 central and effector memory cells. Immunity 35, 583–595 (2011).
Geginat, G., Schenk, S., Skoberne, M., Goebel, W. & Hof, H. A novel approach of direct ex vivo epitope mapping identifies dominant and subdominant CD4 and CD8 T cell epitopes from Listeria monocytogenes. J. Immunol. 166, 1877–1884 (2001).
Rees, W. et al. An inverse relationship between T cell receptor affinity and antigen dose during CD4+ T cell responses in vivo and in vitro. Proc. Natl. Acad. Sci. USA 96, 9781–9786 (1999).
Takeshima, H., Komazaki, S., Nishi, M., Iino, M. & Kangawa, K. Junctophilins: a novel family of junctional membrane complex proteins. Mol. Cell 6, 11–22 (2000).
Fu, X.M., Dai, X., Ding, J. & Zhu, B.T. Pancreas-specific protein disulfide isomerase has a cell type-specific expression in various mouse tissues and is absent in human pancreatic adenocarcinoma cells: implications for its functions. J. Mol. Histol. 40, 189–199 (2009).
Rosette, C. et al. The impact of duration versus extent of TCR occupancy on T cell activation: a revision of the kinetic proofreading model. Immunity 15, 59–70 (2001).
Au-Yeung, B.B. et al. A sharp T-cell antigen receptor signaling threshold for T-cell proliferation. Proc. Natl. Acad. Sci. USA 111, E3679–E3688 (2014).
Klein, L., Kyewski, B., Allen, P.M. & Hogquist, K.A. Positive and negative selection of the T cell repertoire: what thymocytes see (and don't see). Nat. Rev. Immunol. 14, 377–391 (2014).
Tubo, N.J. et al. Single naive CD4+ T cells from a diverse repertoire produce different effector cell types during infection. Cell 153, 785–796 (2013).
Apostolou, I., Sarukhan, A., Klein, L. & von Boehmer, H. Origin of regulatory T cells with known specificity for antigen. Nat. Immunol. 3, 756–763 (2002).
Tanchot, C., Barber, D.L., Chiodetti, L. & Schwartz, R.H. Adaptive tolerance of CD4+ T cells in vivo: multiple thresholds in response to a constant level of antigen presentation. J. Immunol. 167, 2030–2039 (2001).
Kearney, E.R., Pape, K.A., Loh, D.Y. & Jenkins, M.K. Visualization of peptide-specific T cell immunity and peripheral tolerance induction in vivo. Immunity 1, 327–339 (1994).
Lee, H.M., Bautista, J.L., Scott-Browne, J., Mohan, J.F. & Hsieh, C.S. A broad range of self-reactivity drives thymic regulatory T cell selection to limit responses to self. Immunity 37, 475–486 (2012).
PrabhuDas, M. et al. Immune mechanisms at the maternal-fetal interface: perspectives and challenges. Nat. Immunol. 16, 328–334 (2015).
Gupta, R.K. et al. Zfp423 expression identifies committed preadipocytes and localizes to adipose endothelial and perivascular cells. Cell Metab. 15, 230–239 (2012).
Xin, H.B., Deng, K.Y., Rishniw, M., Ji, G. & Kotlikoff, M.I. Smooth muscle expression of Cre recombinase and eGFP in transgenic mice. Physiol. Genomics 10, 211–215 (2002).
Fontenot, J.D. et al. Regulatory T cell lineage specification by the forkhead transcription factor foxp3. Immunity 22, 329–341 (2005).
Shen, A. & Higgins, D.E. The 5′ untranslated region-mediated enhancement of intracellular listeriolysin O production is required for Listeria monocytogenes pathogenicity. Mol. Microbiol. 57, 1460–1473 (2005).
Lauer, P., Chow, M.Y., Loessner, M.J., Portnoy, D.A. & Calendar, R. Construction, characterization, and use of two Listeria monocytogenes site-specific phage integration vectors. J. Bacteriol. 184, 4177–4186 (2002).
Brockstedt, D.G. et al. Listeria-based cancer vaccines that segregate immunogenicity from toxicity. Proc. Natl. Acad. Sci. USA 101, 13832–13837 (2004).
Chu, H.H., Moon, J.J., Kruse, A.C., Pepper, M. & Jenkins, M.K. Negative selection and peptide chemistry determine the size of naive foreign peptide-MHC class II-specific CD4+ T cell populations. J. Immunol. 185, 4705–4713 (2010).
Moon, J.J. et al. Naive CD4+ T cell frequency varies for different epitopes and predicts repertoire diversity and response magnitude. Immunity 27, 203–213 (2007).
Malhotra, D. et al. Transcriptional profiling of stroma from inflamed and resting lymph nodes defines immunological hallmarks. Nat. Immunol. 13, 499–510 (2012).
Berzins, S.P., Boyd, R.L. & Miller, J.F. The role of the thymus and recent thymic migrants in the maintenance of the adult peripheral lymphocyte pool. J. Exp. Med. 187, 1839–1848 (1998).
de Hoon, M.J., Imoto, S., Nolan, J. & Miyano, S. Open source clustering software. Bioinformatics 20, 1453–1454 (2004).
Eisen, M.B., Spellman, P.T., Brown, P.O. & Botstein, D. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. USA 95, 14863–14868 (1998).
Acknowledgements
We thank D. Mueller for reviewing the manuscript; J. Walter and O. Rainwater for technical assistance; P. Lauer (and Aduro Biotech) for the Lm-eGFP strain PL1113. Supported by the US National Institutes of Health (P01 AI35296 to M.K.J. and K.A.H.; F32 AI114050 to D.M.; T32 GM008244 and F30 DK093242 to R.W.N.; T32 AI07313 to J.L.L.; K99 AI114884 to Y.J.L.), the Wallin Neuroscience Discovery Fund (to H.T.O. and M.K.J.) and the Juvenile Diabetes Research Foundation (2-2011-662 to B.T.F.).
Author information
Authors and Affiliations
Contributions
D.M. designed the study, did experiments, analyzed data, and wrote the manuscript, J.L.L., T.D., Y.J.L., W.E.P., J.V.L., R.W.N. and M.S.A. did experiments, B.T.F., H.T.O. and K.A.H. provided discussions; and M.K.J. designed the study, analyzed data and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
Supplementary Figure 1 Gating strategy for thymic epithelial cells and dendritic cells.
(a) Contour plots depicting the initial gating strategy for analysis of eGFP and eYFP expression by EpCAM+ thymic epithelial cells and dendritic cells. Briefly, thymuses were enzymatically digested to produce single cell suspensions, which were enriched for EpCAM+ and CD11c+ cells. Doublets were excluded. Numbers indicate the percent of cells in each gate.
(b) Contour plots showing the gating strategy for EpCAM+ thymic epithelial cells, proceeding from singlet gate 2 (a). EpCAM+ CD45.2− stromal cells were gated for further analysis by excluding CD90.2+ T cells, dead cells, CD19+ B cells, macrophages and dendritic cells (CD11b+ and CD11c+). Remaining cells were analyzed further in Figure 7. Numbers indicate the percent of cells in the gate.
(c) Contour plots showing the gating strategy for thymic dendritic cells, proceeding from singlet gate 2 (a). CD11c+ cells were gated and dead cells, CD90.2+ T cells, CD11chiCD11bhi eosinophils, CD19+ B cells, and autofluorescent macrophages (shaded, blue gate) were then excluded. Remaining CD11c+CD11blo–int cells were analyzed further in Figure 7. Numbers indicate the percent of cells carried forward for analysis.
Supplementary information
Supplementary Text and Figures
Supplementary Figure 1 and Supplementary Table 1 (PDF 568 kb)
Rights and permissions
About this article
Cite this article
Malhotra, D., Linehan, J., Dileepan, T. et al. Tolerance is established in polyclonal CD4+ T cells by distinct mechanisms, according to self-peptide expression patterns. Nat Immunol 17, 187–195 (2016). https://doi.org/10.1038/ni.3327
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ni.3327
This article is cited by
-
A guide to thymic selection of T cells
Nature Reviews Immunology (2024)
-
Type 2 cytokines in the thymus activate Sirpα+ dendritic cells to promote clonal deletion
Nature Immunology (2022)
-
Factors that influence the thymic selection of CD8αα intraepithelial lymphocytes
Mucosal Immunology (2021)
-
Inducible de novo expression of neoantigens in tumor cells and mice
Nature Biotechnology (2021)
-
Cellular and molecular mechanisms breaking immune tolerance in inborn errors of immunity
Cellular & Molecular Immunology (2021)